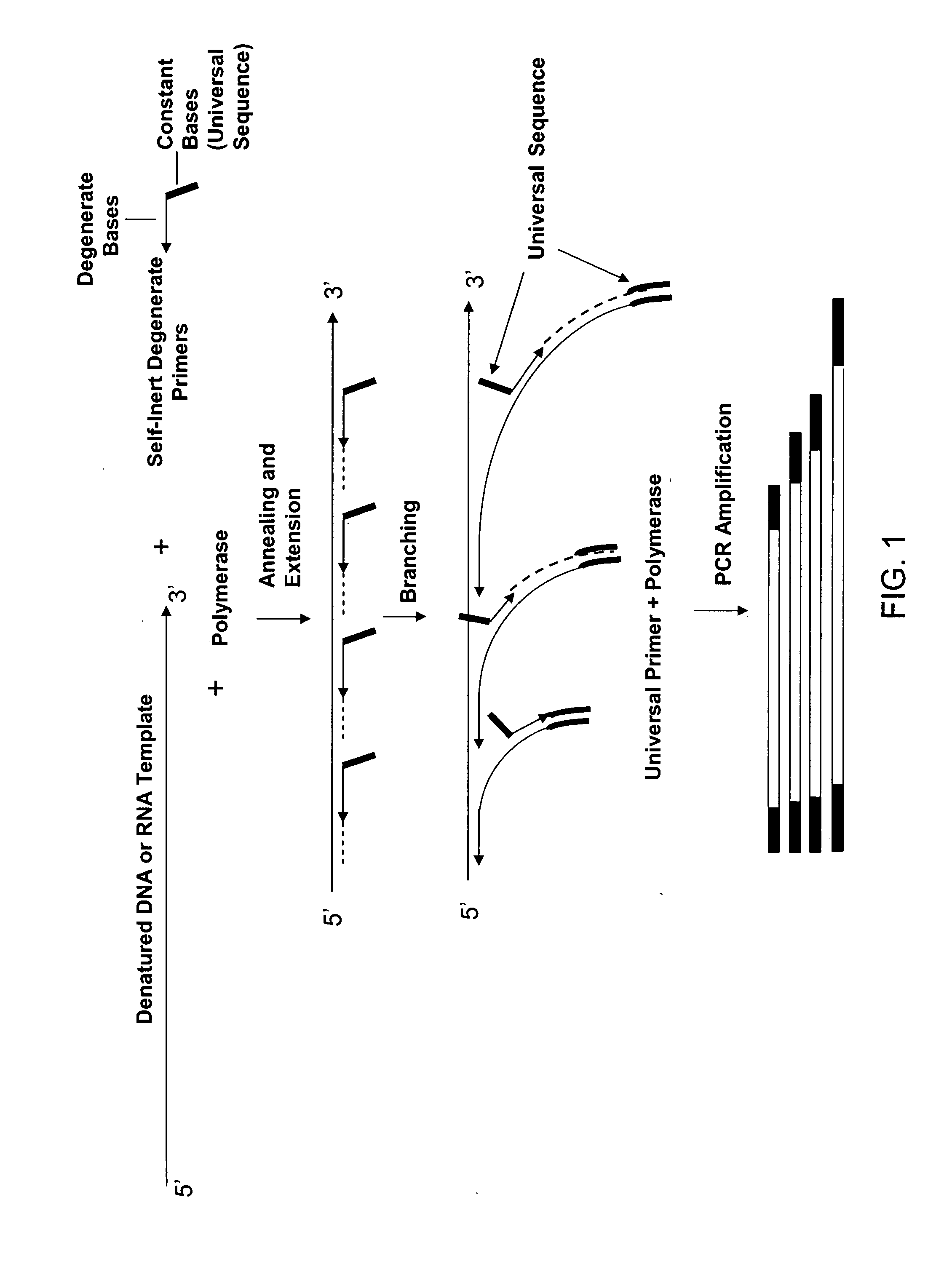
Amplification and analysis of whole genome and whole transcriptome libraries generated by a DNA polymerization process
a polymerization and dna technology, applied in the field of amplify, can solve the problems of inability to amplify single stranded, short, or fragmented dna and rna molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Design of Degenerate Pyrimidine Primers and Analysis of Self-Priming and Extension
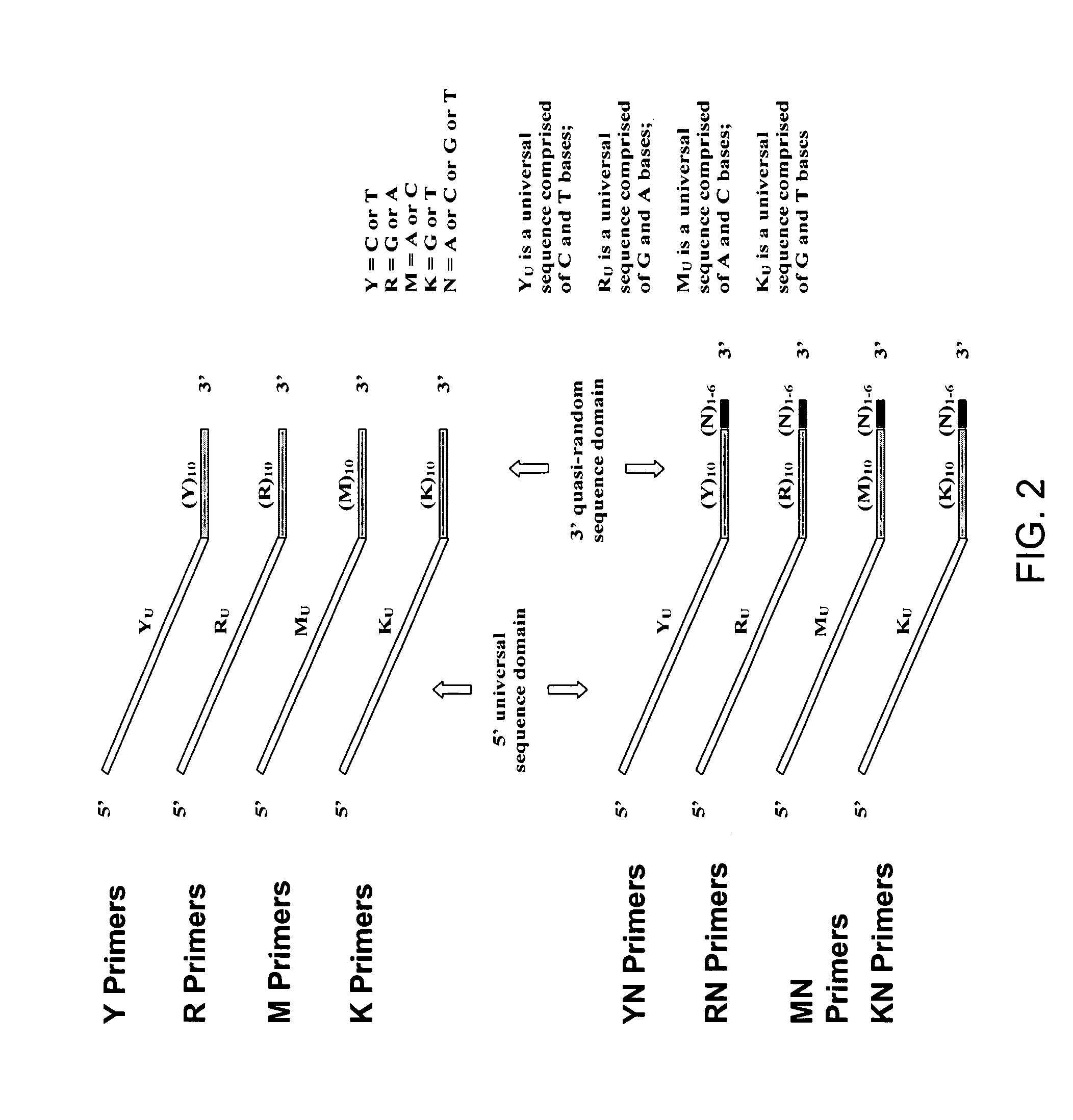
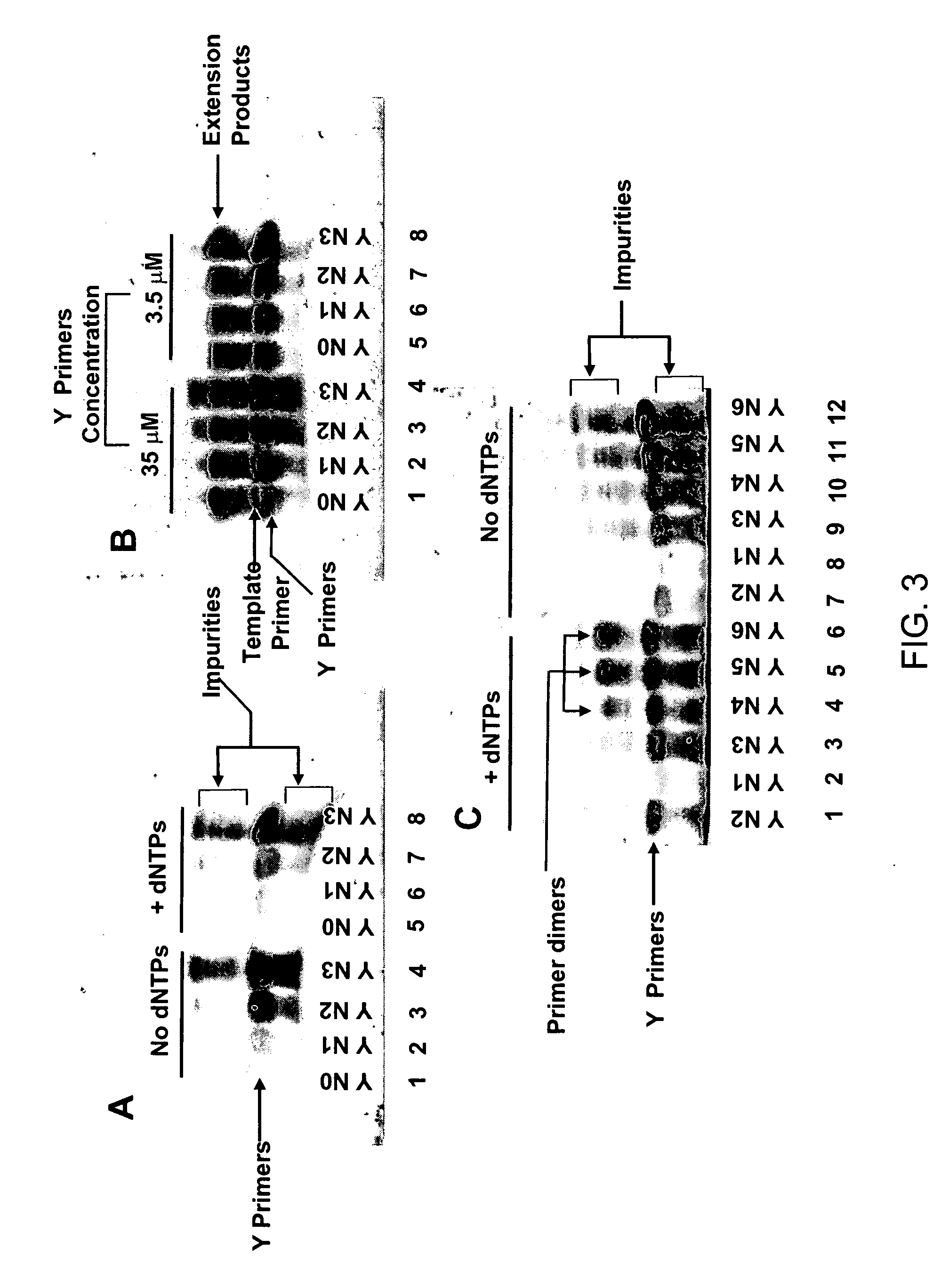
[0254] Pyrimidine primers comprising a constant 18 base sequence, followed by 10 random pyrimidines and between 0 and 6 completely random bases at the 3′ end (Table III, primers 1-7), are compared for their ability to self prime and to extend a model template oligonucleotide.
TABLE IIIOLIGONUCLEOTIDE SEQUENCESNoCodeSequence 5′-3′ *1.YCCTTTCTCTCCCTTCTCTYYYYYYYYYY(SEQ ID NO:11)2.YNCCTTTCTCTCCCTTCTCTYYYYYYYYYYN(SEQ ID NO:12)3.Y(N)2CCTTTCTCTCCCTTCTCTYYYYYYYYYYNN(SEQ ID NO:13)4.Y(N)3CCTTTCTCTCCCTTCTCTYYYYYYYYYYNNN(SEQ ID NO:14)5.Y(N)4CCTTTCTCTCCCTTCTCTYYYYYYYYYYNNNN(SEQ ID NO:15)6.Y(N)5CCTTTCTCTCCCTTCTCTYYYYYYYYYYNNNNN(SEQ ID NO:16)7.Y(N)6CCTTTCTCTCCCTTCTCTYYYYYYYYYYNNNNNN(SEQ ID NO:17)8.YuCCTTTCTCTCCCTTCTCT(SEQ ID NO:18)9.TemplateGTAATACGACTCACTATAGGRRRRRRRRRR(SEQ ID NO:19)10.R(N)2AGAGAAGGGAGAGAAAGGRRRRRRRRRRNN(SEQ ID NO:20)11.RuAGAGAAGGGAGAGAAAGG(SEQ ID NO:21)12.M(N)2CCAAACACACCCAACACAMMMMMMMMMMNN(SEQ I...
example 2
Comparison of Different Degenerate Pyrimidine Primers Used in the Library Synthesis with Klenow Exo− Fragment of DNA Polymerase-I and Subsequent Whole Genome Amplification
[0256] Human lymphocyte genomic DNA isolated by standard procedures was randomly fragmented in TE buffer to an average size of 1.5 Kb using the Hydro Shear™ device (Gene Machines; Palo Alto, Calif.). The reaction mixture contained 50 ng of fragmented DNA in 1× EcoPol buffer (NEB), 200 μM of each dNTP, 360 ng of Single Stranded DNA Binding Protein (USB), 500 nM of known YU primer (Table III, primer 8), and 1 μM of degenerate pyrimidine primers with 0 to 6 random 3′ bases (Table III, primers 1-7) or 1 μM of T7 primer with six random N bases at the 3′ end (Table III, T7(N)6 primer 16,) in a final volume of 25 μl. After a denaturing step of 2 min at 95° C., the samples were cooled to 16° C., and the reaction was initiated by adding 5 units of Klenow enzyme that lacks 3′-5′ exonuclease activity (NEB). WGA library synth...
example 3
Whole Genome Amplification of Thermally Fragmented Genomic DNA Converted into an Amplifiable DNA Library Using Klenow Exo− Fragment of DNA Polymerase-I or Sequenase Version-2 and Degenerate Primers Y(N)2
[0260] Human lymphocyte genomic DNA isolated by standard procedures was randomly fragmented in TE-L buffer (10 mM Tris, 0.1 mM EDTA, pH 7.5) by heating at 95° C. for 5 min. The reaction mixture contained 100 ng of thermally fragmented DNA in 1× EcoPol buffer (NEB) or 1× Sequenase buffer (USB), 200 μM of each dNTP, 360 ng of Single Stranded DNA Binding Protein (USB), 200 nM of known YU primer (Table III, primer 8), and 500 nM of degenerate Y(N)2 primer (Table III, primer 3) in a final volume of 25 μl. After a denaturing step of 2 min at 95° C., the samples were cooled to 16° C., and the reaction initiated by adding 2.5 units or 6.5 units of Klenow Exo− polymerase (NEB) or Sequenase version 2 (USB), respectively. WGA library synthesis was carried out in a three-step protocol for 10 mi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com