Method for detecting embryonic chromosome abnormality by virtue of blastochyle free DNA
A chromosomal abnormality and blastocoel fluid technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., to achieve the effects of low probability, simple operation, and convenient material acquisition.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1 Mouse blastocoel fluid free DNA detection
[0060] (1) Collection of blastocoel fluid
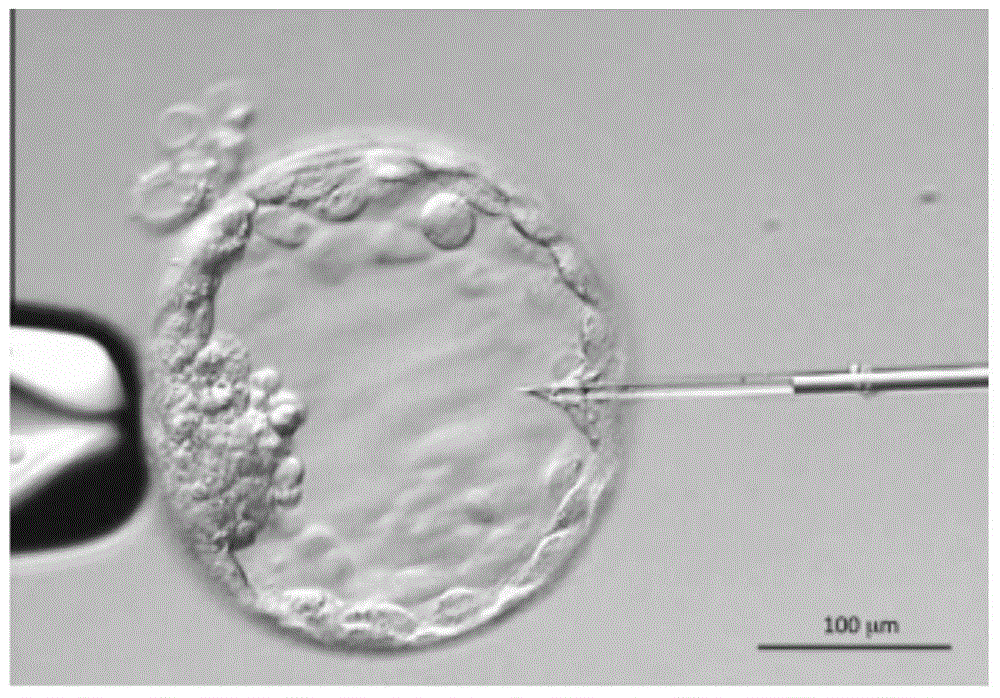
[0061] Acquisition and processing of mouse blastocoel fluid: the acquisition of mouse blastocoel fluid DNA and the acquisition of mouse single cells are the same mouse embryo, so as to facilitate the comparison of test results. In the micromanipulation laboratory, samples were taken by micropuncture technique at room temperature. In the process of aspirating blastocoel fluid DNA, avoid cytoplasm contamination and damage to trophoblast cells. The DNA sampling process of mouse blastocoel fluid is as follows: figure 1 shown.
[0062] (2) Target gene and primer design
[0063] Two genes, GAPH and TBCD, were used as detection genes, and primers were designed for quantitative analysis and detection. The sizes of the amplified fragments of GAPDH and TBCD were 100bp and 120bp.
[0064] Amplification primer sequences are as follows.
[0065] GAPDH:
[0066] P1 TGTCTCACC TTAT...
Embodiment 2
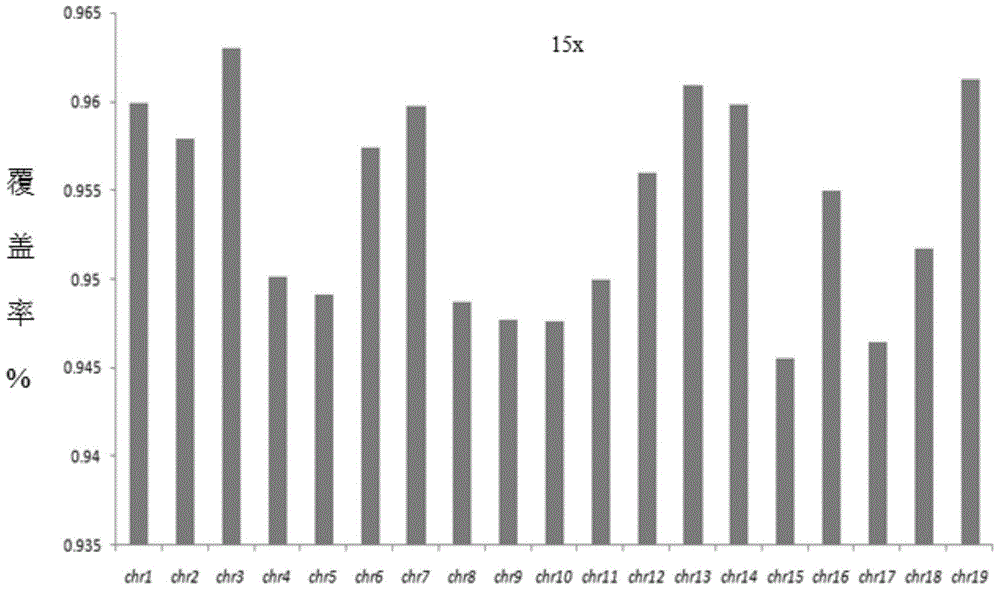
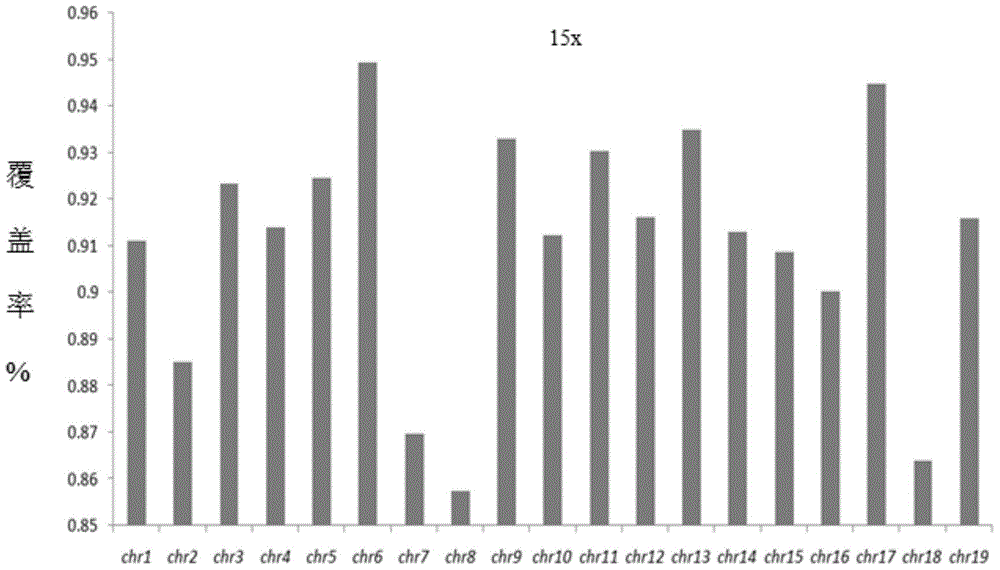
[0077] Comparison of mouse genome sequencing coverage (mouse embryo single cell and mouse blastocoel fluid DNA)
[0078] (1) Single cell isolation and blastocoel fluid DNA acquisition and processing:
[0079] Pick 10 mouse embryo single-cell samples and 10 mouse blastocoel fluid samples, and each mouse embryo single-cell sample and mouse blastocoel fluid sample correspond one-to-one, and come from the same mouse embryo.
[0080] a. Isolation and processing of mouse embryo single cells: under a microscope, single cells were isolated by laser drilling technology, and the isolated single cells were placed in a 200 μl PCR tube containing less than 2 μl of PBS, and water was added to a final volume of 9 μl. Add 1 μl of freshly prepared cell lysis buffer to the PCR tube and mix well. The resulting mixture was incubated: 50°C for 1h, 99°C for 4min, 15°C hold, cooled on ice, and centrifuged.
[0081] b. Acquisition and processing of mouse blastocoel fluid: the acquisition of mouse b...
Embodiment 3
[0098] Chromosomal Abnormality Analysis of Mouse Genome Sequencing (Mouse Embryo Single Cell and Mouse Blastocoel Fluid DNA)
[0099] (1) The purpose of this experiment is to analyze that blastocoel fluid can be used for chromosomal abnormality analysis instead of single-cell samples. Two gene knockout mice from our laboratory were selected, one of which was chromosomal deletion and the other was chromosomal duplication. Each mouse was subjected to blastocoel fluid acquisition and embryonic single cell isolation. After the genomic DNA was amplified and the amplified product was qualified, fragmentation processing and library construction were performed. The experimental procedure was the same as in Example 2.
[0100] (2) On-machine sequencing. The quality inspection of the samples constructed in the above experiments was carried out, and the quality inspection was qualified, and the Ion Proton system was used for whole genome sequencing.
[0101] (3) Biological information ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com