Method for simultaneously completing gene locus, chromosome and linkage analysis
A gene locus and linkage analysis technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of inability to perform batch detection, allele tripping, and high detection costs, and is conducive to popularization and application, short working cycle, simple and fast operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
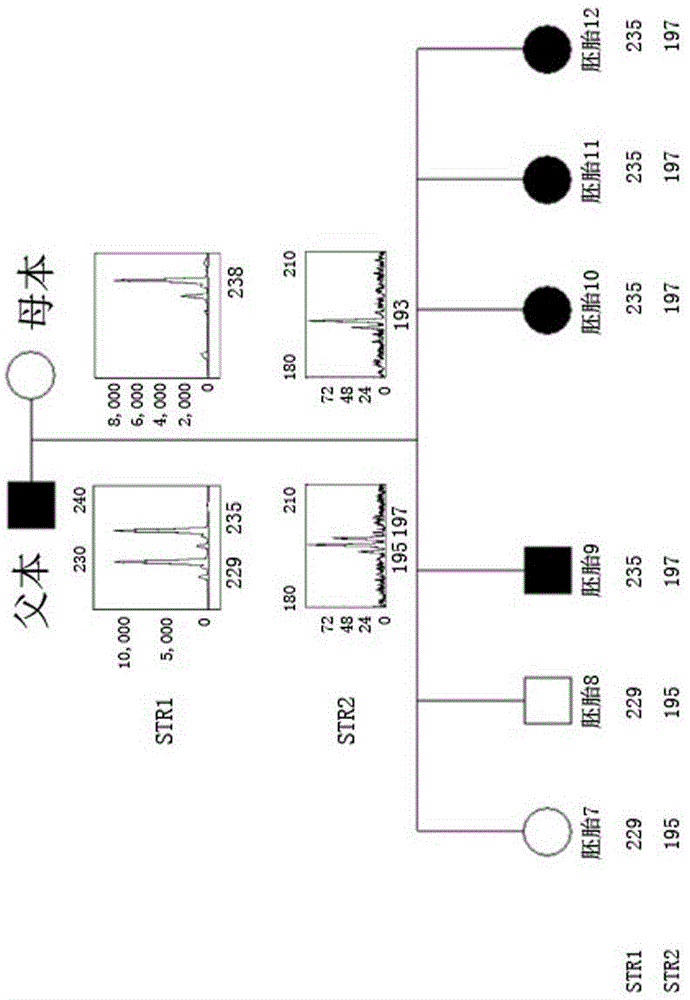
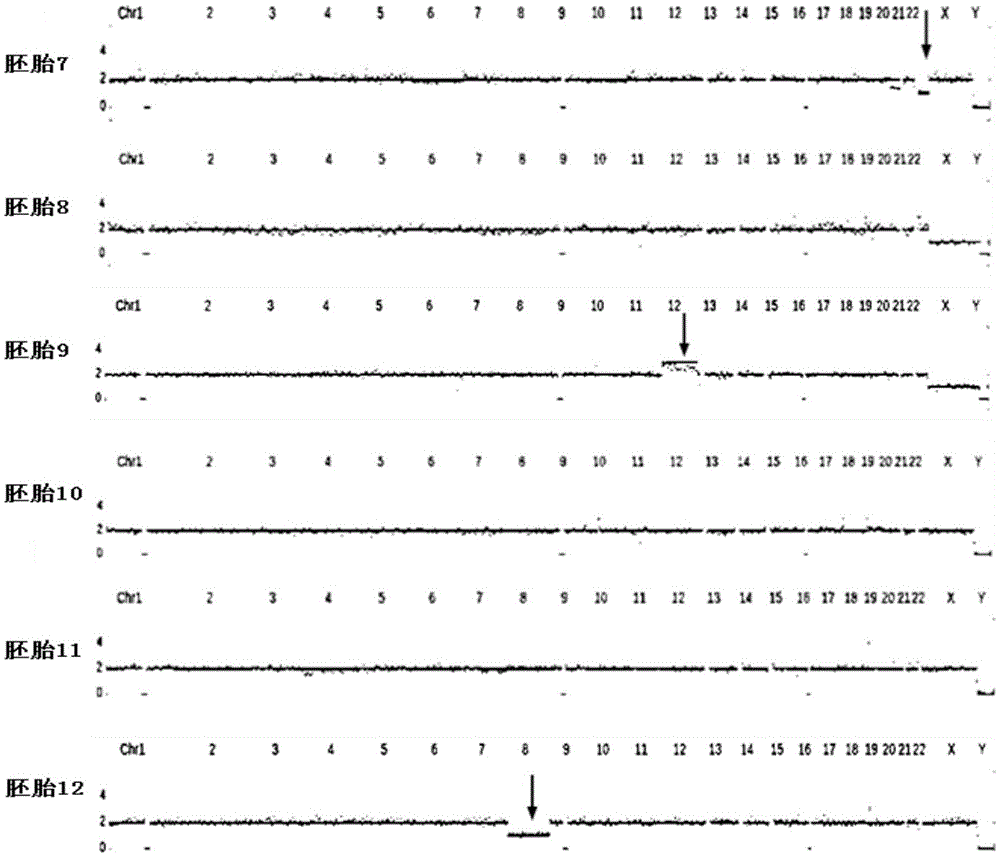
[0069] An autosomal dominant genetic disease model, the causative gene is A. In this family, both the father and the father of the father are carriers, respectively carrying the Mutation mutation site (c.233delC). After analysis, embryos 9, 10, 11 and 12 were pathogenic embryos of paternal mutation carriers, and embryos 7 and 8 did not carry the mutation site.
[0070] 1. Take embryonic single cells:
[0071] 1.1 Fertilization of eggs
[0072] Microinjection of single sperm (ICSI) to eggs in the MII stage, put the eggs in (G-MOPS) operating solution, transfer to the platform of the micromanipulator, and perform micromanipulation.
[0073] 1.2 In vitro culture of embryos
[0074] The fertilized embryos were cultured in G1 culture medium (Vitrolife) or Gm culture medium (Global) for about 72 hours to the 5-8 cell stage, laser-drilled on the zona pellucida, and transferred to the balanced G2 culture medium (Vitrolife ) or Gm culture medium (Global) to continue culturing to bl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com