Method for conducting SNP-haplotype analysis by means of multiplex PCR technology
A haplotype analysis and haplotype technology, applied in the field of molecular biology and bioinformatics, can solve the problems of long distance, easy recombination, high detection cost and long cycle.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
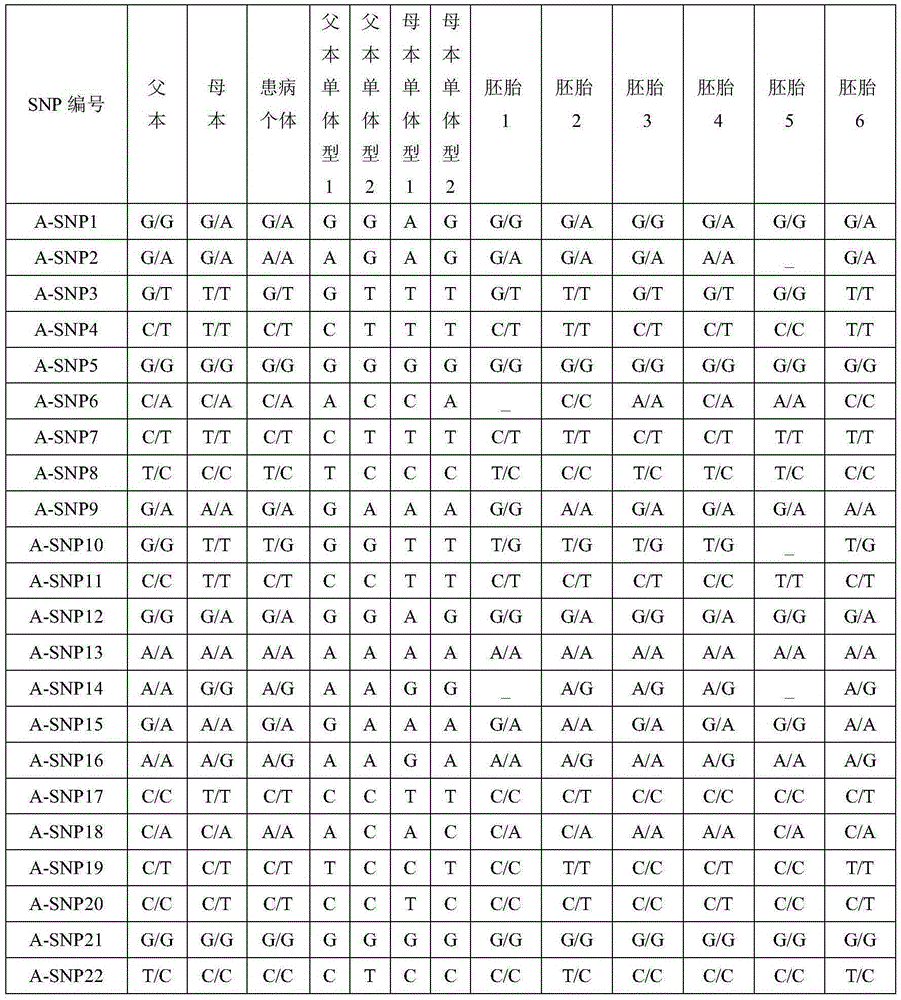
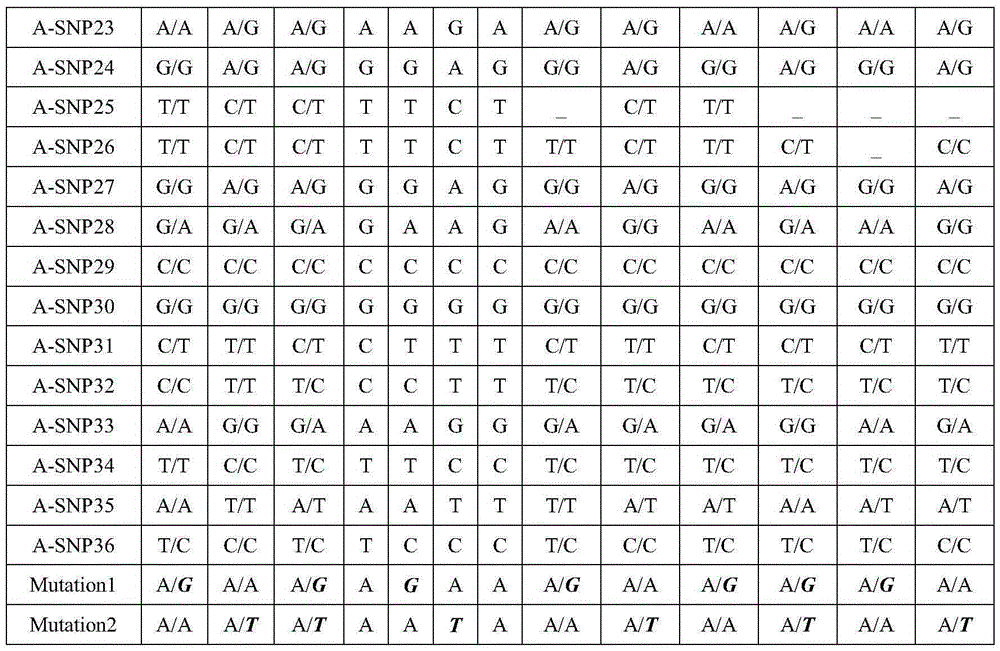
[0050] The SNP-haplotype analysis was carried out according to the above steps, and the specific results are shown in Table 1. An autosomal recessive genetic disease model, the causative gene is A. In this family, both father and mother are carriers, carrying Mutation1 and Mutation2 mutation sites respectively.
[0051] After analysis, for A-SNP1, the paternal allelic type is G / G, the maternal allelic type is G / A, and the affected individual's allelic type is G / A, indicating that the base of this site in the maternal A is linked with the pathogenic site in the same haplotype and is inherited to the affected individual. This SNP site is a distinguishing SNP; the alleles of embryos 2, 4 and 6 all have base A, indicating that this All three embryos carried maternal pathogenic loci. According to the principle of Mendelian inheritance, determine whether the embryo carries the disease-causing gene locus: Embryos 1, 3, and 5 are carriers of the paternal mutation, embryos 2 and 6 ar...
Embodiment 2
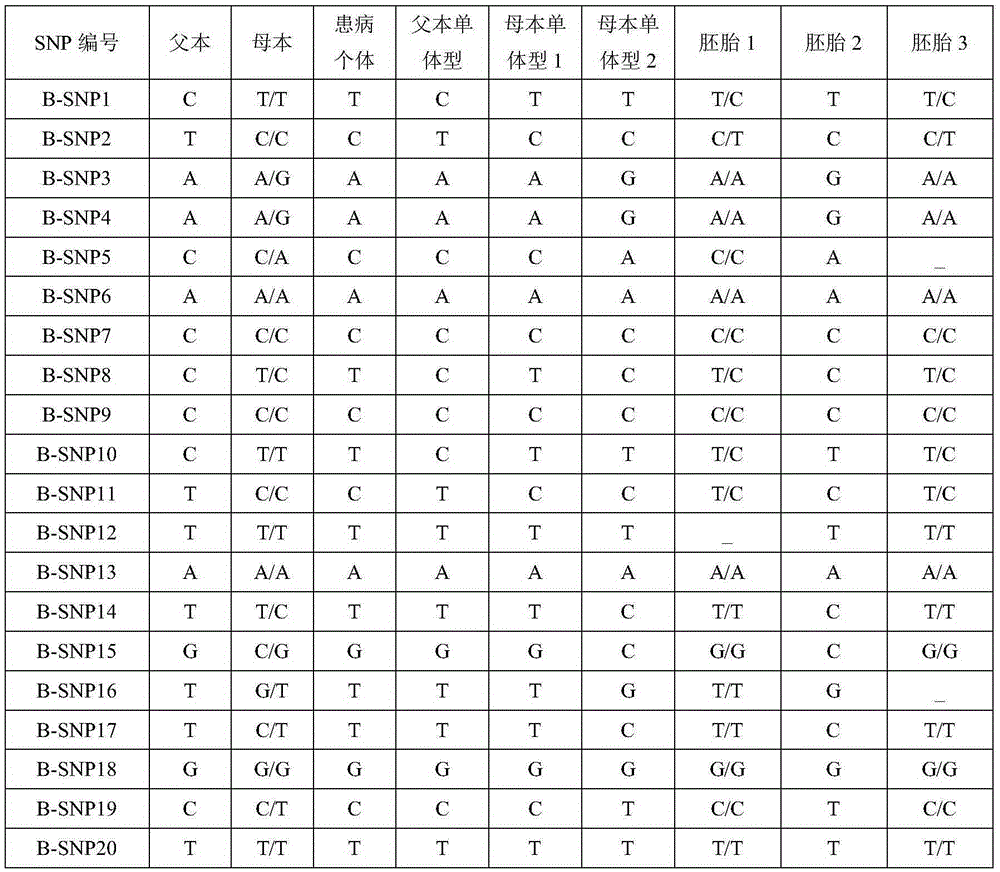
[0053] The SNP-haplotype analysis was carried out according to the above steps, and the specific results are shown in Table 2. An X-linked recessive genetic disease model, the causative gene is B. In this family, the female parent is a carrier of the B gene Mutation3 mutation site. According to the above analysis, it can be seen that embryo 2 does not carry the pathogenic mutation, and embryos 1 and 3 are carriers of the pathogenic mutation.
[0054] Table 1A Partial SNP genotypes in the gene region
[0055]
[0056]
[0057] Table 2B Partial SNP genotypes in the gene region
[0058]
[0059]
[0060] Notes:
[0061] 1. "_" in the table indicates that the corresponding SNP data cannot be obtained (no data coverage or low depth);
[0062] 2. Bold italics indicate pathogenic mutations;
[0063] 3. The haplotype 1 of the father and mother in Table 1 indicates the haplotype of the pathogenic mutation, and the maternal haplotype 1 in Table 2 indicates the haplotype...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com