Method for analyzing transcription and translation activity of functional genes of microbial community
A technology of microbial community and functional genes, applied in the field of analyzing the transcription and translation activities of functional genes of microbial communities, can solve the problem of inconsistency, the inability to understand the mechanism of transcriptional regulation and maintaining functional homeostasis of functional genes of microbial communities, and cannot reveal the community Problems such as gene dynamic expression and gene regulation process, to achieve the effect of increasing integrity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
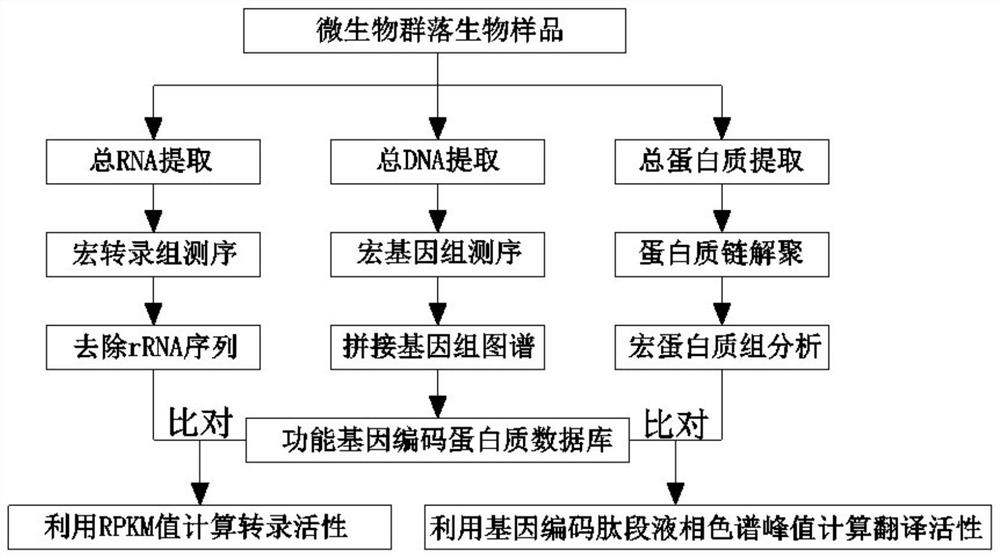
[0034] In this embodiment, the analysis of gene transcription and translation expression activity in the process of carbon metabolism and nitrogen metabolism of denitrification microbial flora mainly includes the following steps:
[0035] 1. Extract the total DNA sample, total RNA sample and total protein sample of the microbial community
[0036] First, the same denitrification microbial flora was selected, and then these flora were cultured for 60 days at different concentrations of culture substrates, and then the cultures were taken as microbial samples for analysis. Among them, microbial sample one (R 1 ) in the culture medium of ammonia nitrogen and nitrite nitrogen concentration are 300mg / L, microbial sample two (R 2 ) The concentration of ammonia nitrogen and nitrite nitrogen in the culture medium is 300mg / L. Second, extract a total DNA sample, a total RNA sample, and a total protein sample from the two microbial samples. Among them, the PowerSoil DNA Isolation Kit ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com