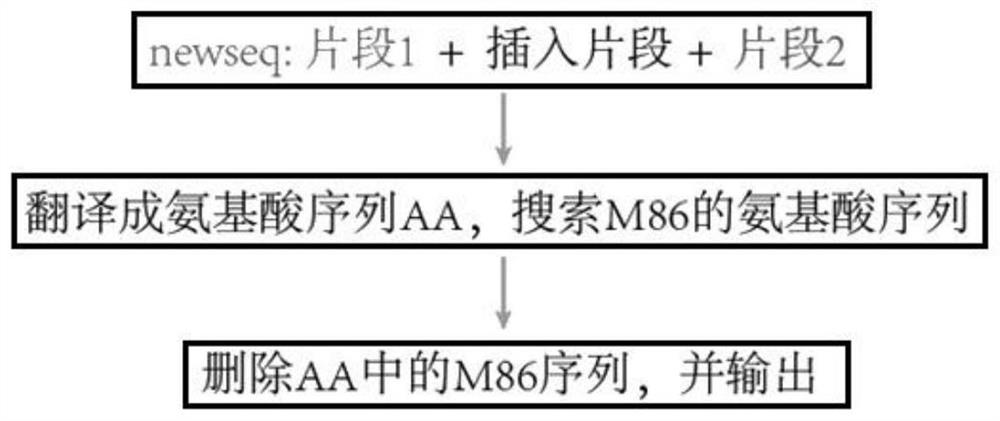
Method for screening splitting sites and application thereof
A site, amino acid technology, applied in the field of screening split sites, can solve problems such as lack of characteristics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] can refer to image 3 and Figure 4 In this example, the above-mentioned screening method was used to verify the polypeptide breakage site of Escherichia coli antigenic protein (Im7-6).
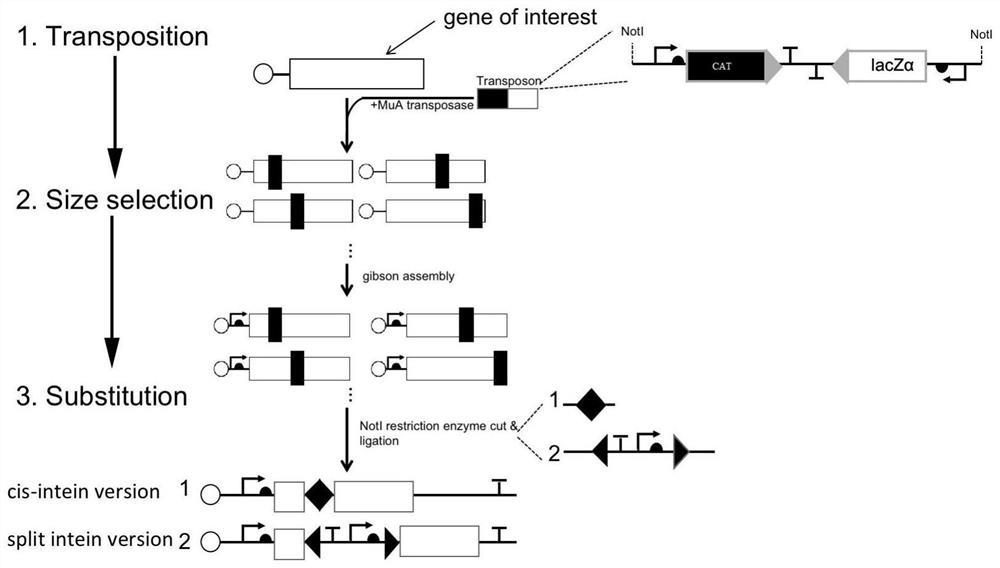
[0042] like image 3 shown, image 3 It is the same experimental principle as the above-mentioned scientific document "A systematic approach to insertingsplit inteins for Boolean logic gate engineering and basal activityreduction", that is, the method of randomly inserting gene fragments by mini-Mu transposon mediated by the phage transposition mechanism. The specific operations are as follows:
[0043] 1. Use the transposase MuA to carry out the transposition experiment on the target gene fragment, and insert the transposon into the target fragment randomly, and the principle ensures that only one transposon fragment is inserted into each target gene fragment. The transposon fragment is a complete expression line with promoter, terminator and other elements and expressing chloramph...
Embodiment 2
[0052] In this example, the above-mentioned screening method was used to verify the two polypeptide cleavage sites of the Cas9 protein.
[0053] The steps 1-5 are roughly the same as those in Example 1, the difference is that in step 3, "replace the transposon fragment with the expression intein Ssp DnaBM86" by the restriction enzyme digestion Replace the transposon fragment with the N-terminal, terminator and promoter transcription elements of the split intein Ssp DnaBM86 and the gene fragment of the C-terminal of the split intein Ssp DnaBM86 (corresponding to image 3 3. The split intein version method in Substitution)"
[0054] like Figure 7 Shown is a schematic representation of the nucleotide sequence of the intein Ssp DnaBM86 with restriction sites and additional bases added to prevent frameshift mutations. Ditto Figure 4 said, Figure 7 GCGGCCGC in the two boxes 1 is the nucleotide sequence of the restriction enzyme cleavage site, the C bases in the two boxes 2 ar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com