Genome editing method based on CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat) system and application thereof
A genome editing and genome sequence technology, applied in the field of genome editing, can solve the problems of unable to control the expression amount synchronously, unable to construct inducible expression or spatiotemporal specific expression genome editing technology, etc. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] inPTG-Cas9 for multi-site editing in rice
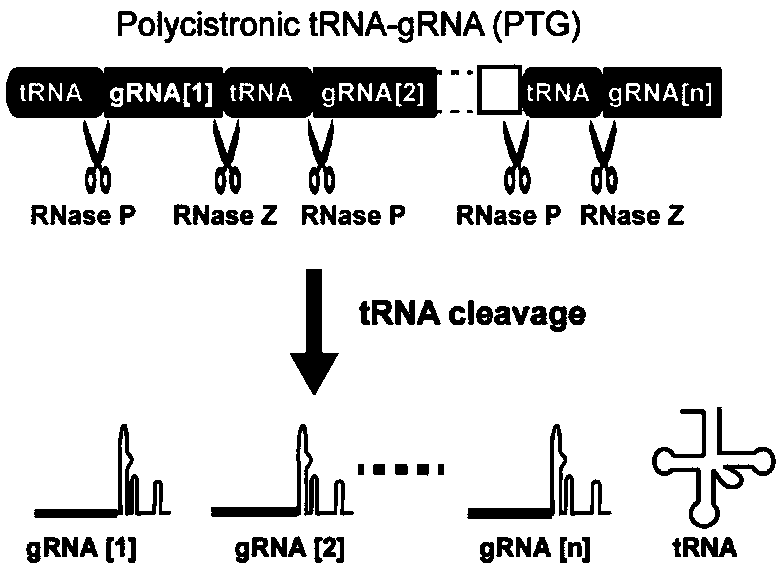
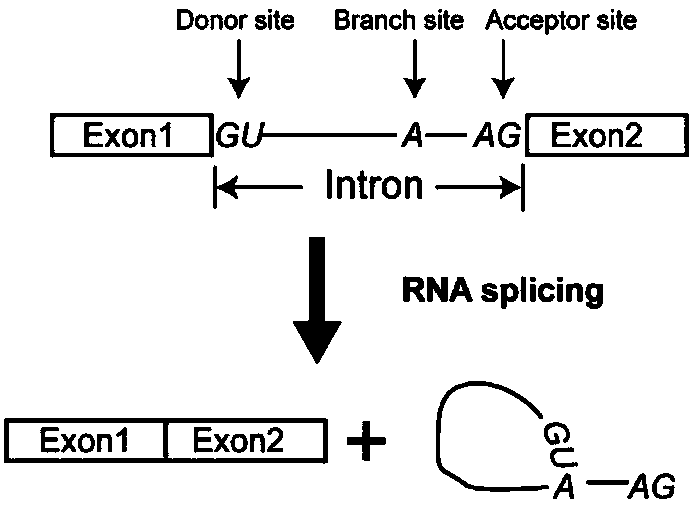
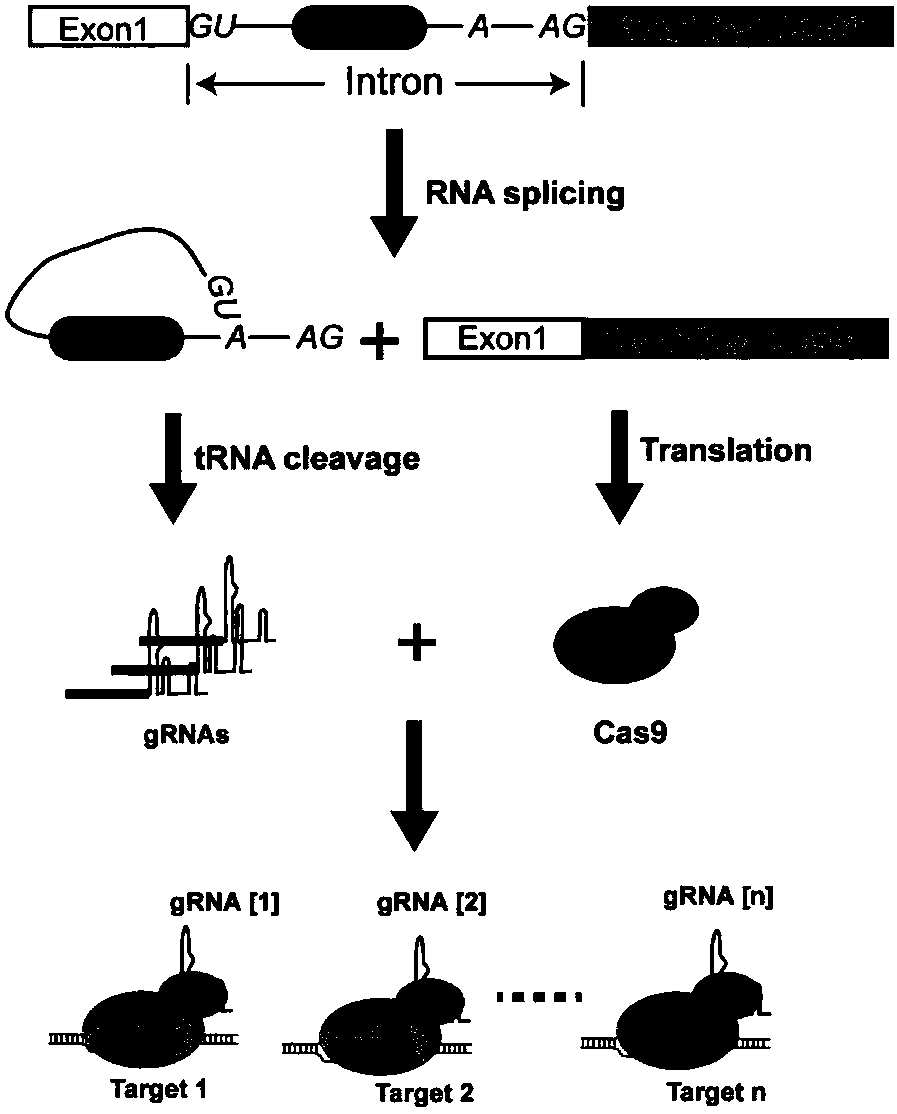
[0070] The design is as follows: multiple tRNA-gRNA tandem units (polycistronic tRNA-gRNA, PTG for short) are placed in the intron of the coding gene. The intron containing inPTG and the exon encoding Cas9 are fused into a gene, and such fusion gene can express Cas9 protein and gRNA simultaneously. When the inPTG-Cas9 system is expressed in plants, the PTG fragment in the spliced intron is recognized and processed by RNase P and Z, and a single gRNA is released, while the splicing of the intron does not affect the expression of the Cas9 gene. Transcription and translation, such an inPTG-Cas9 fusion gene can efficiently edit multiple target sites at the same time.
[0071] 1. Carrier Description
[0072] In order to realize the expression of inPTG, we modified the rice UBI10 gene, and inserted the PTG sequence structure into the intron of UBI 10 (such as image 3 shown). There is a 962bp intron (sequence such as SEQ ID No...
Embodiment 2
[0161] Example 2 Verification of inPTG-Cas9 editing efficiency in rice
[0162] We further verified the editing efficiency of inPTG-Cas9 in rice plants. The pRGEB33-inPTG3 / 6 / 7 / 10 vectors were transformed into Zhonghua 11 rice plants by the method of Agrobacterium-mediated transformation.
[0163] pRGEB33-inPTG10 is used to knock out the PDS gene. When the PDS gene is knocked out, the rice plants will appear albino. Among the transgenic plants, albino seedlings appeared in 12 plants among the 27 T0 generation transgenic plants that knocked out PDS (see Figure 13 ), the results showed that inPTG10-Cas9 has high editing efficiency.
[0164] Using PCR / RE method to detect MPK1, MPK5, PDS editing in transgenic rice. Use the CTAB method to extract the genomic DNA of the transgenic rice leaves, use the genomic DNA of the wild-type rice as a control, use primers (see primer sequence table 7) to amplify the genomic DNA containing the target site, and use restriction enzymes to diges...
Embodiment 3
[0170] Example 3 The intron of inPTG can be placed in different positions of the gene
[0171] The design of inPTG-Cas9 uses the structure of rice endogenous UBI10 gene to express Cas9 and gRNA in the intron. Since the intron position of the gene is not fixed, inPTG can be expressed at different positions of the recombinant gene. In order to verify our hypothesis, we constructed a new structure to insert inPTG into the 3'-UTR region of the Cas9 gene (UBI10p::Cas9-inPTG). At the same time, tRNA (inPTG-tRNA) is added after the last gRNA of PTG to improve the efficiency of cutting the last target site from the intron.
[0172] 1. Carrier Description
[0173] On the basis of the vector pRGEB33, the intron with the BsaI restriction site was replaced with the endogenous intron of UBI10 (SEQ ID: 4), and two BsaI enzymes were inserted into the 3'-UTR region of the recombinant Cas9 gene The intron of the cleavage site (SEQ ID: 9) is used for PTG cloning, and the vector is named pRGE...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com