Compression and clustering-based batch protein homology search method
A search method and protein technology, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve the problems of time-consuming, large memory space and storage space consumption, etc., to achieve the reduction of repeated sequence alignment and no gaps Extended Time Effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0028] The specific implementation manners of the present invention will be described in detail below in conjunction with the technical solutions and accompanying drawings.
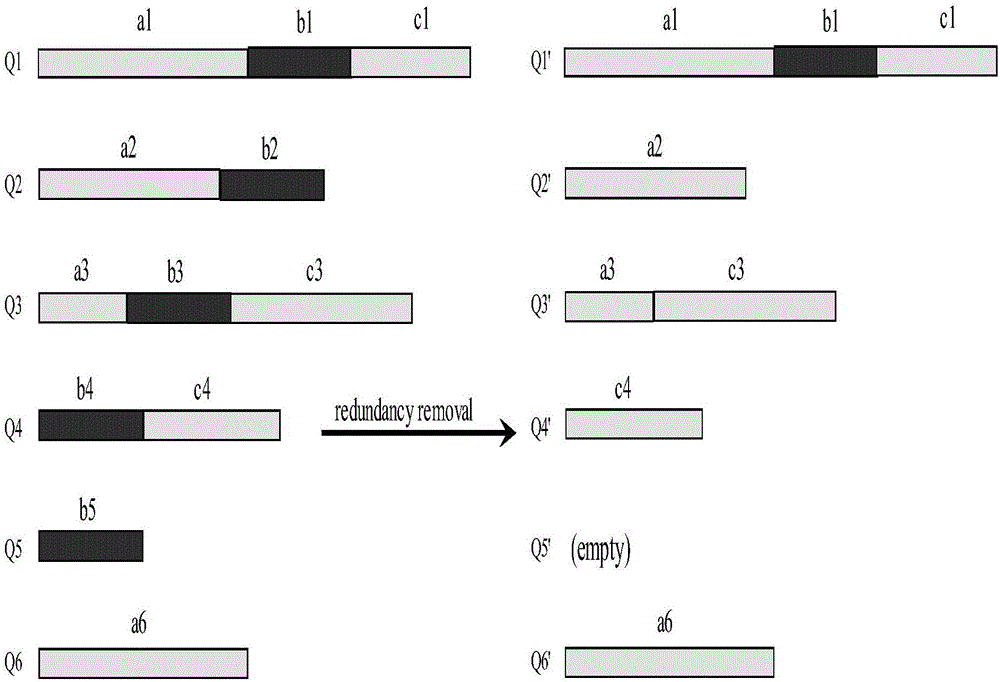
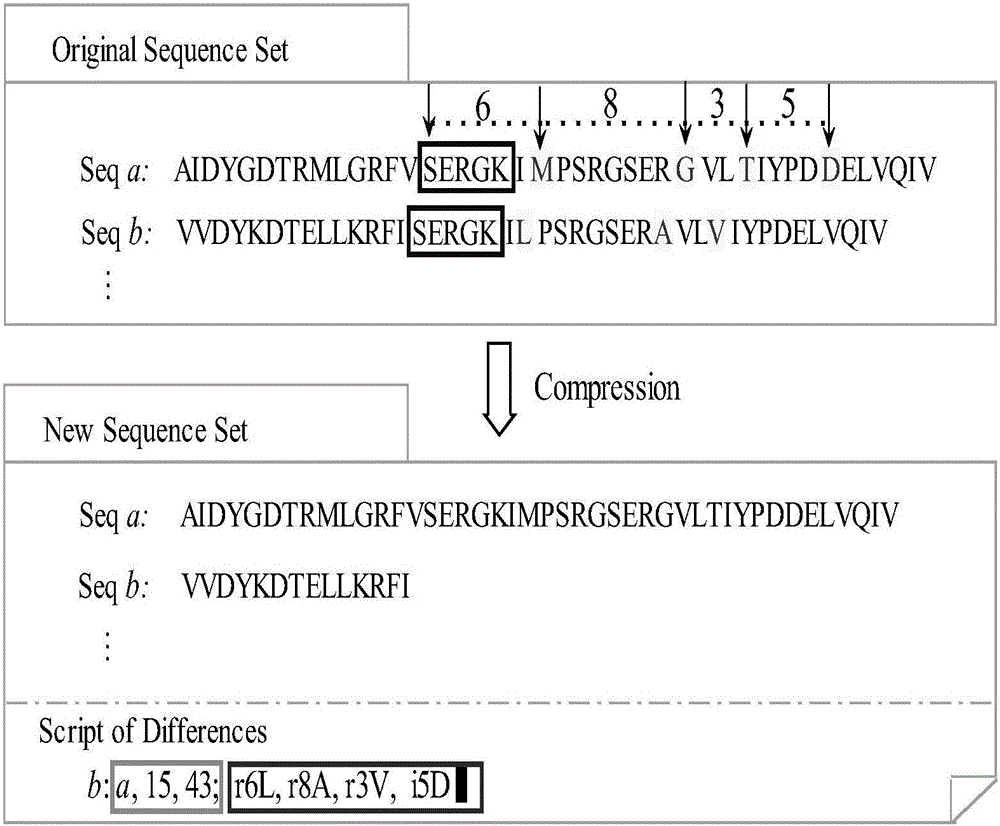
[0029] The access mainly includes three stages of protein sequence compression, clustering and batch search.
[0030] 1. The specific steps of compressing query sequences and protein database sequences in offline state are as follows:
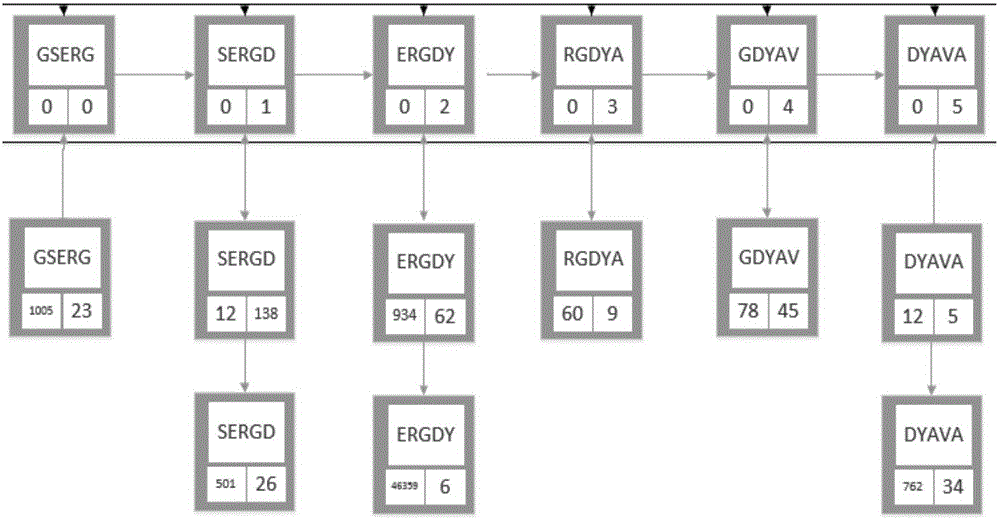
[0031] 1) Scan a protein sequence from left to right to create a key-entry mapping set, such as figure 1 As shown, in each key-entry mapping of the mapping set, the key is a protein sequence fragment composed of 5 amino acids, and the entry includes three attributes: sequence number, starting amino acid position, and pointer to the next sequence.
[0032] 2) Scan a new protein sequence from left to right, and the new protein sequence fragment is also composed of 4-6 amino acids; apply the Needleman-Wunsch algorithm to perform similarity between each new protein sequence f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com