A 3D protein structure prediction method based on Monte Carlo local dithering and fragment assembly
A prediction method and technology of three-dimensional structure, applied in special data processing applications, instruments, electrical and digital data processing, etc., can solve problems such as error convergence, falling into a minimum value, and difficulty in obtaining a globally optimal and stable concept, so as to reduce the search space. Complexity, the effect of improving the convergence ability
Active Publication Date: 2016-06-22
ZHEJIANG UNIV OF TECH
View PDF0 Cites 0 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
[0007] In order to overcome the shortcomings of using a single modern evolutionary algorithm to predict the three-dimensional structure of proteins in the prior art, it is easy to fall into the minimum value, it is difficult to obtain the global optimal stable concept, the prediction efficiency is low, and the shortcomings of wrong convergence
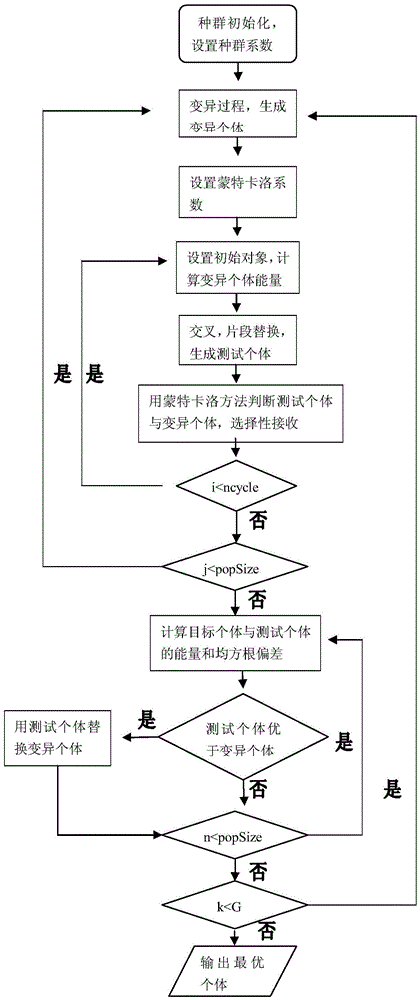
The present invention combines the Monte Carlo statistical simulation method with the existing known knowledge base, and proposes a protein three-dimensional structure prediction method based on Monte Carlo local shaking and fragment assembly, which effectively obtains the local optimal stable concept, and has high prediction efficiency , the convergence accuracy is better, making up for the existing search space is too complex and the above defects
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
example
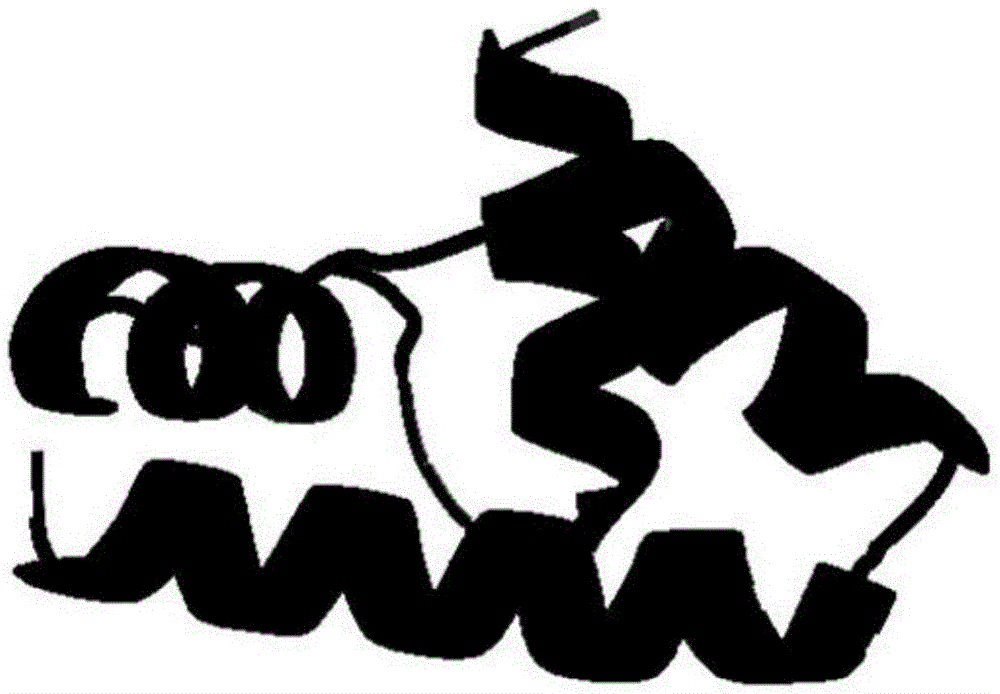
[0058] Example: Taking the protein numbered 1GYZ as an example, it is a protein molecule composed of 60 amino acids, and the amino acid sequence is:
[0059] WIARINAAVRAYGLNYSTFINGLKKAGIELDRKILADMAVRDPQAFEQV
[0060] VNKVKEALQVQ.
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
The invention discloses a method for predicting a protein three-dimensional structure based on Monte Carlo local shaking and fragment assembly. The method comprises the following steps that firstly, according to the difficult problem that search space of protein high-dimensional conformation space is complex, the effectiveness of fragment replacement is judged under a Rosetta force field model through the Monte Carlo statistical method according to a protein database configuration fragment bank; under a differential evolution group algorithm framework, the complexity of the search space is reduced through fragment assembly, meanwhile, false fragment assembly is removed through the Monte Carlo statistical method, and the conformation search space is gradually reduced through the diversity of an evolutionary algorithm, and therefore the searching efficiency is improved; meanwhile, a module with coarseness is adopted, a side chain is ignored, and cost of a search is effectively reduced. The method for predicting the protein three-dimensional structure based on Monte Carlo local shaking and fragment assembly can effectively obtain optimal local stable conformation and is high in predicting efficiency and good in convergence correctness.
Description
technical field [0001] The invention relates to the technical field of protein three-dimensional structure prediction in bioinformatics, in particular to a protein three-dimensional structure prediction method, which belongs to the application of modern intelligent optimization methods to protein three-dimensional structure prediction. Background technique [0002] Bioinformatics reveals the biological mysteries of large and complex biological data through the comprehensive use of biology, computer science and information technology. It is a hot spot of current research. Bioinformatics research results have been widely used in sequence alignment, protein alignment, gene recognition analysis, molecular evolution, sequence contig assembly, genetic code, drug design, biological systems, protein structure prediction, etc. Among them, protein structure prediction is an important branch in the field of bioinformatics. Anfinsen, the famous Nobel laureate in chemistry, showed that...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Patents(China)
IPC IPC(8): G06F19/16
Inventor 张贵军陈先跑周晓根秦传庆张贝金明洁刘玉栋
Owner ZHEJIANG UNIV OF TECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com