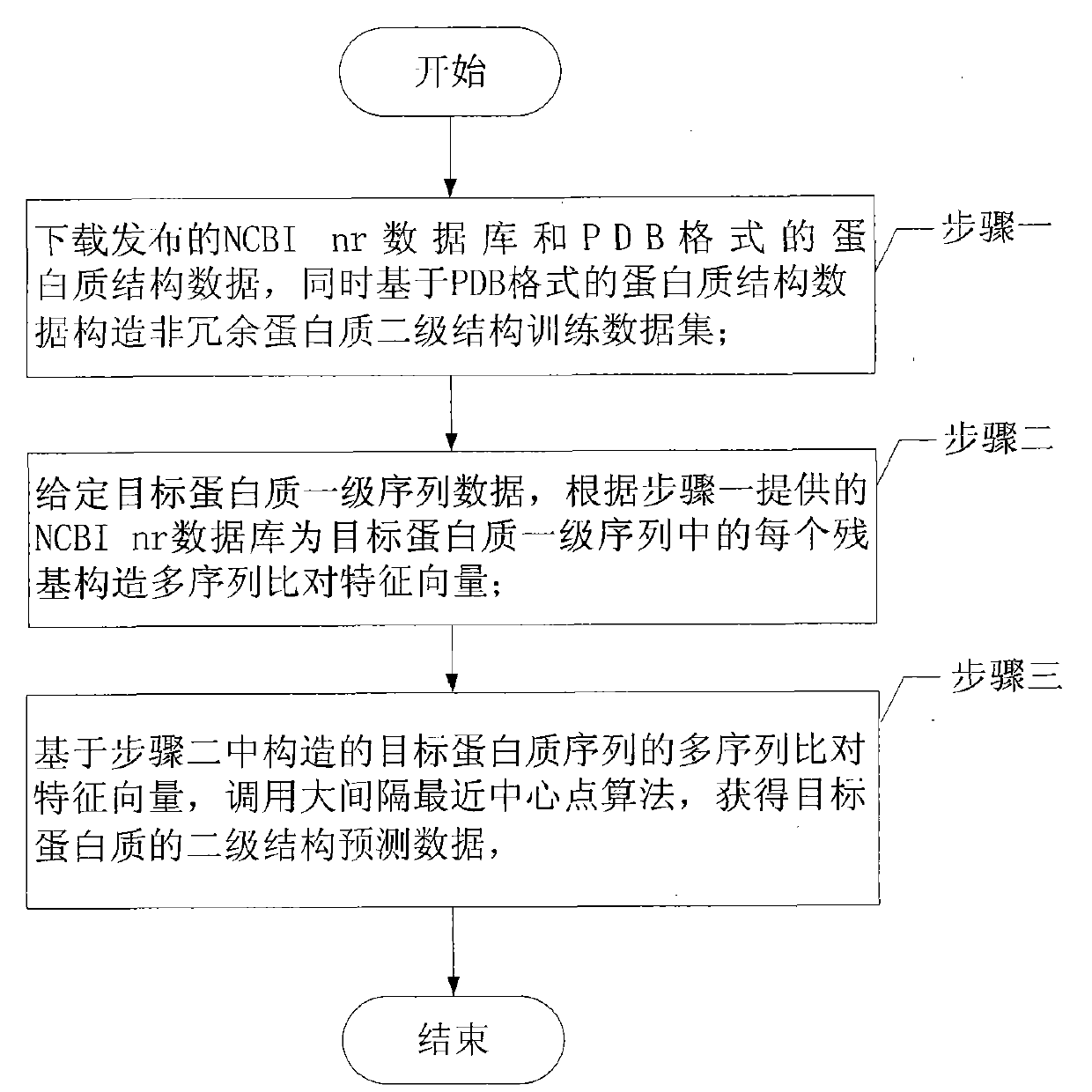
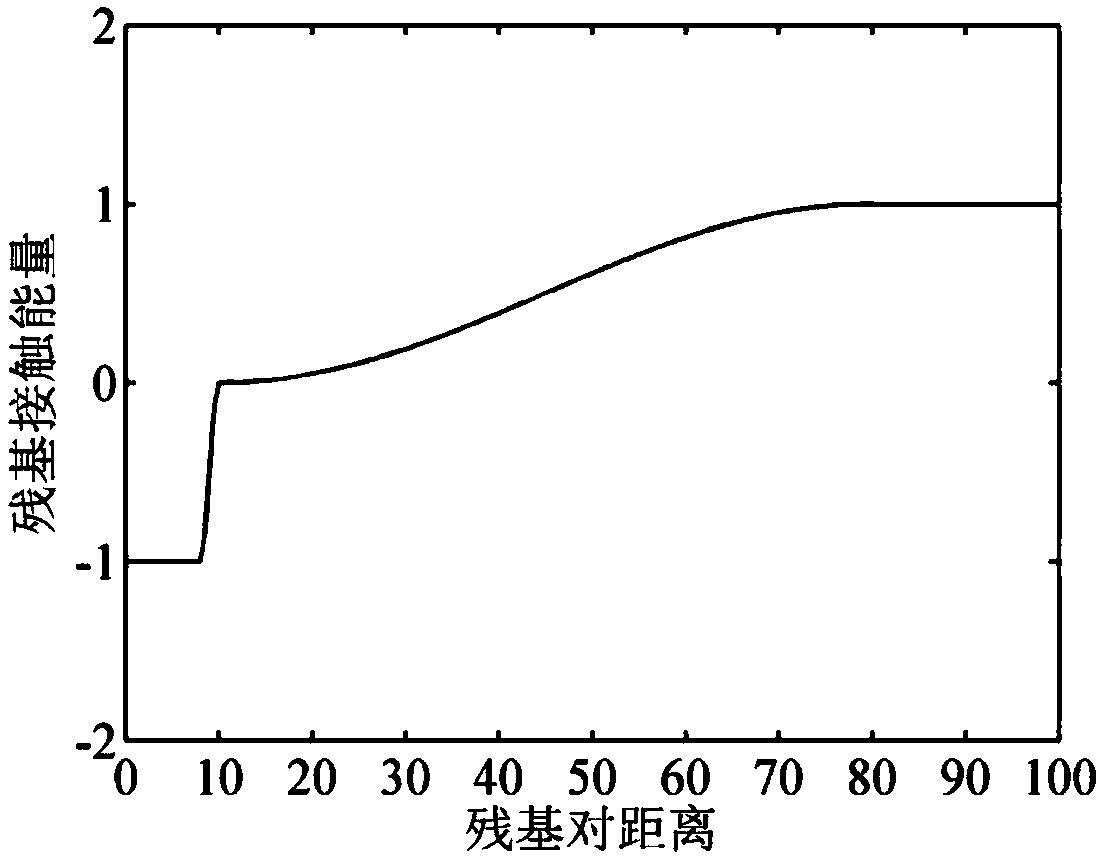
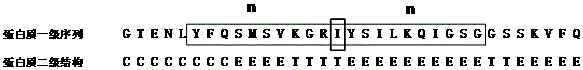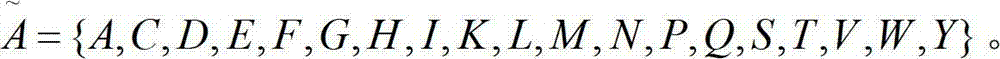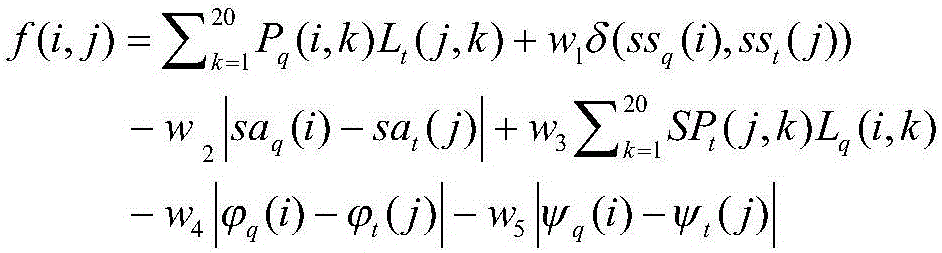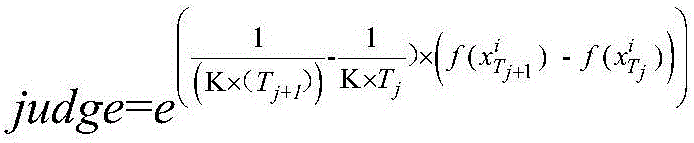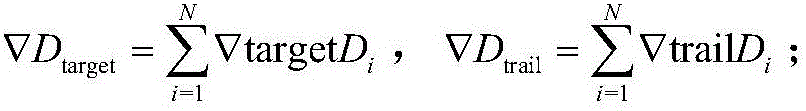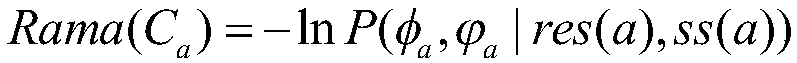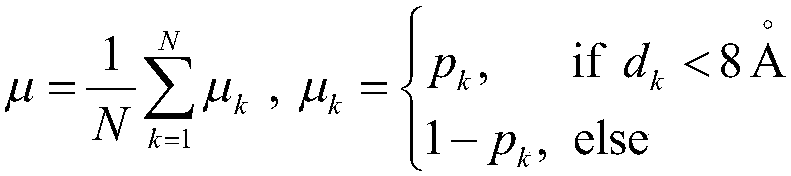Patents
Literature
198 results about "Protein structure prediction" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
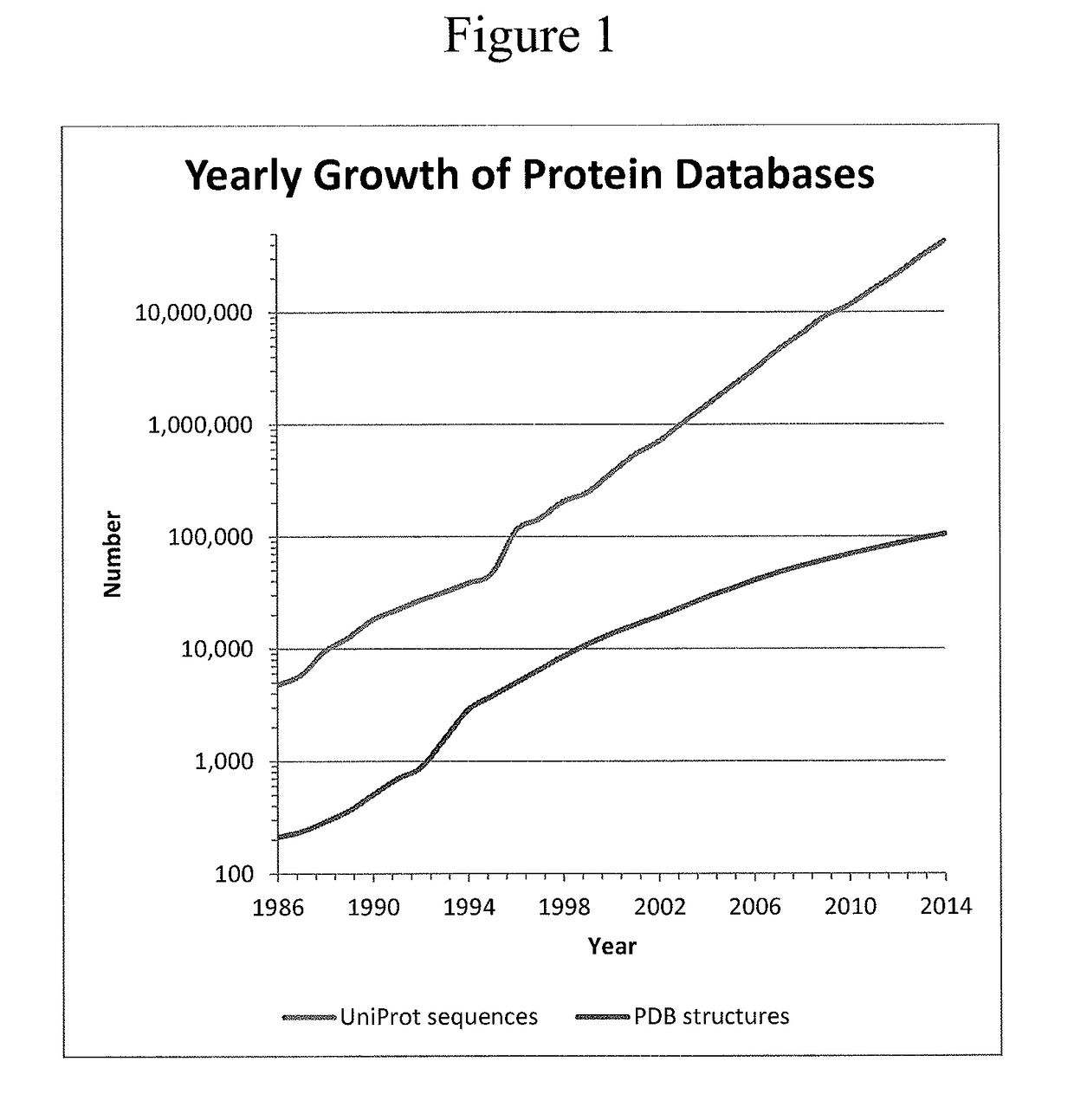
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its folding and its secondary and tertiary structure from its primary structure. Structure prediction is fundamentally different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by bioinformatics and theoretical chemistry; it is highly important in medicine (for example, in drug design) and biotechnology (for example, in the design of novel enzymes). Every two years, the performance of current methods is assessed in the CASP experiment (Critical Assessment of Techniques for Protein Structure Prediction). A continuous evaluation of protein structure prediction web servers is performed by the community project CAMEO3D.
Abstract convex lower-bound estimation based protein structure prediction method
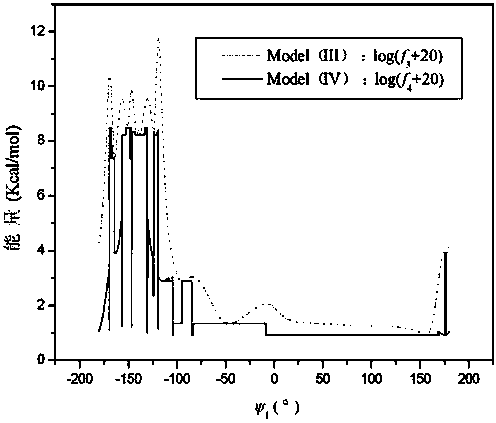
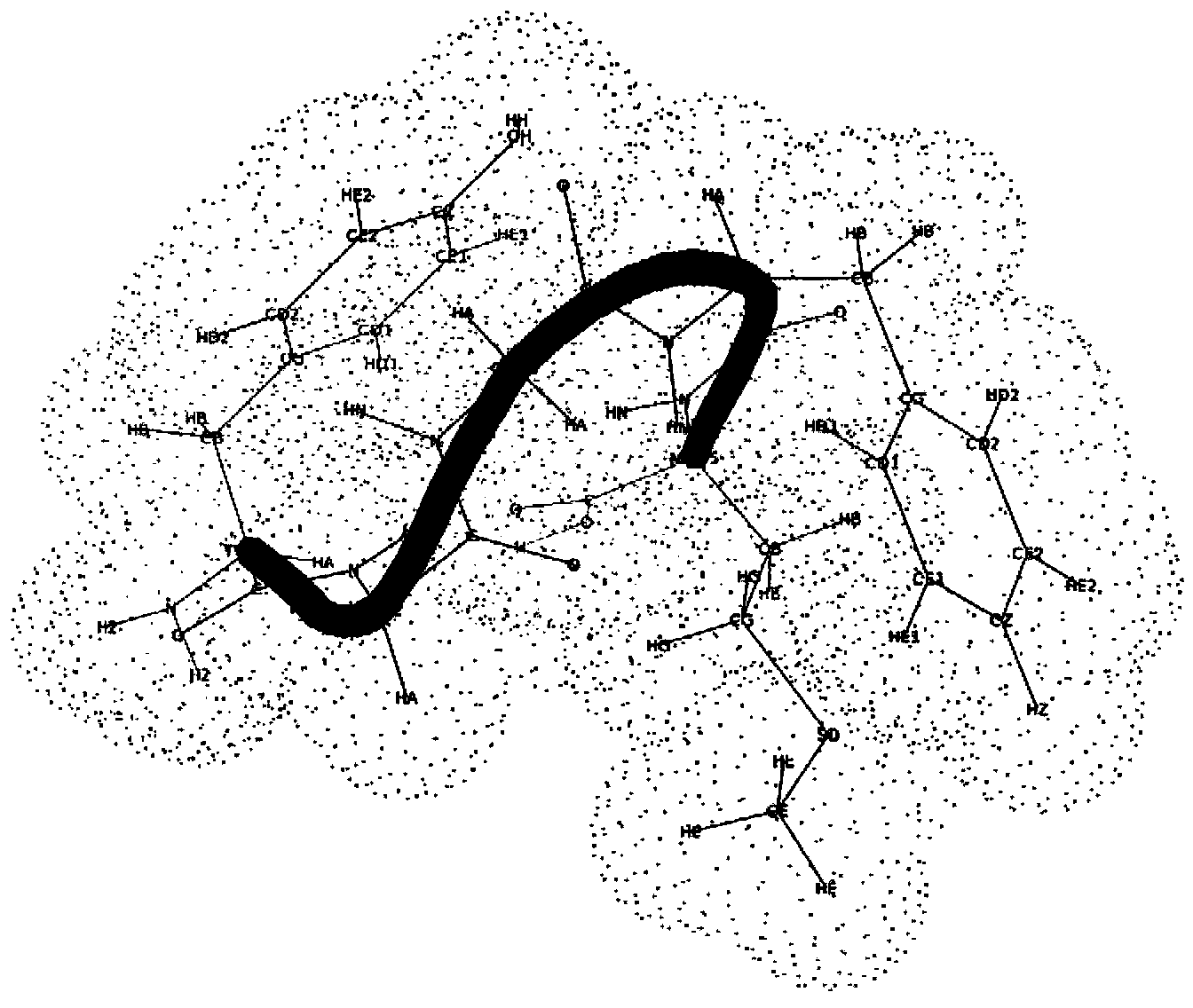
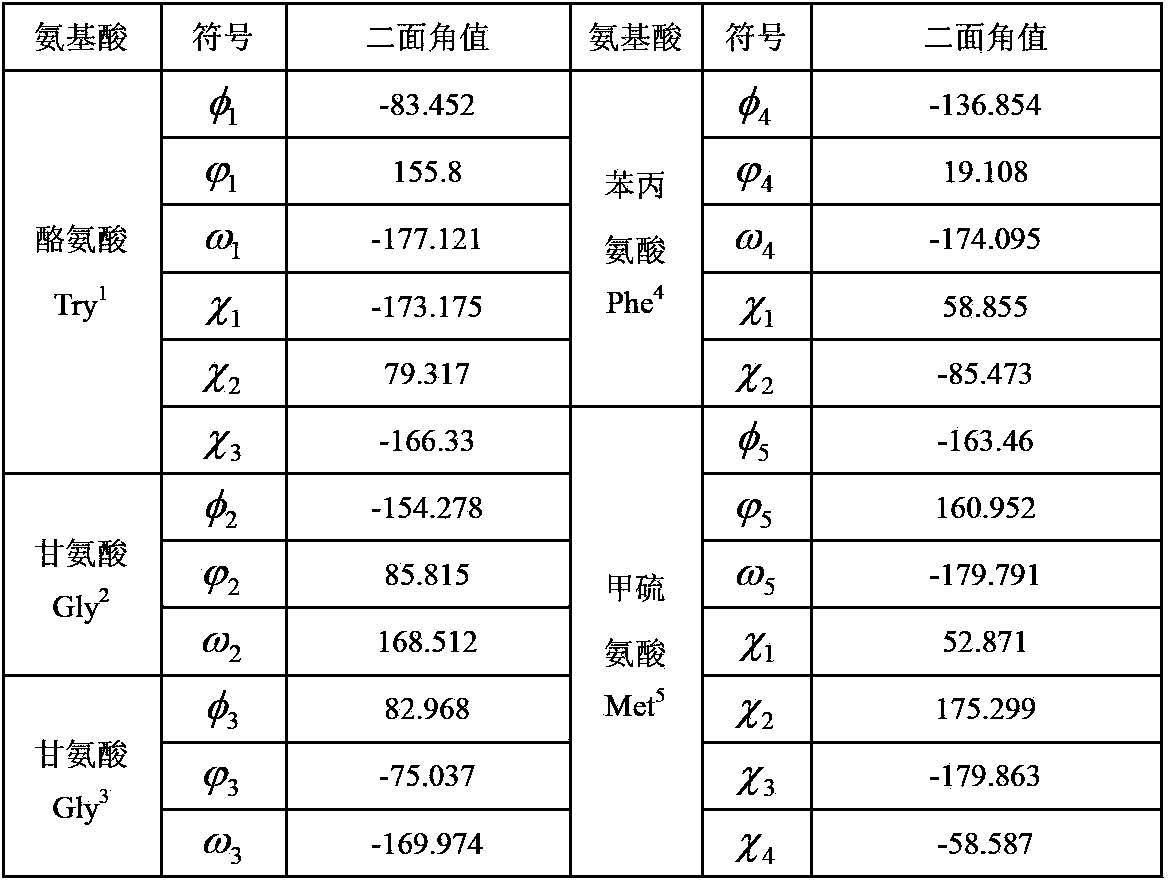
Disclosed is an abstract convex lower-bound estimation based protein structure prediction method. The method includes: firstly, aiming for high-dimensional conformational spatial sampling problems for proteins, adopting a series of transform methods to transform an ECEPP / 3 force field model into an increasing radial convex function in unit simple constraint conditions; secondly, based on an abstract convex theory, proving and analyzing to give out a supporting hyperplane set of the increasing radial convex function; thirdly, constructing a lower-bound underestimate supporting plane on the basis of population minimization conformation subdifferential knowledge under a differential evolution population algorithm framework; fourthly, by the aid of a quick underestimate supporting plane extreme point enumeration method, gradually decreasing a conformational sampling space to improve sampling efficiency; fifthly, utilizing the lower-bound underestimate supporting plane for quickly and cheaply estimating an energy value of an original potential model to effectively decrease evaluation times of a potential model objective function; finally, verifying effectiveness of the method by methionine-enkephalin (TYR1-GLY2-GLY3-PHE4-MET5) conformational spatial optimization examples. The abstract convex lower-bound estimation based protein structure prediction method is high in reliability, low in complexity and high in computation efficiency.
Owner:ZHEJIANG UNIV OF TECH
Hierarchical multi-label classification method for protein function prediction
ActiveCN106126972ASolve the multi-label problemAchieve forecastBiostatisticsProteomicsData setProtein function prediction
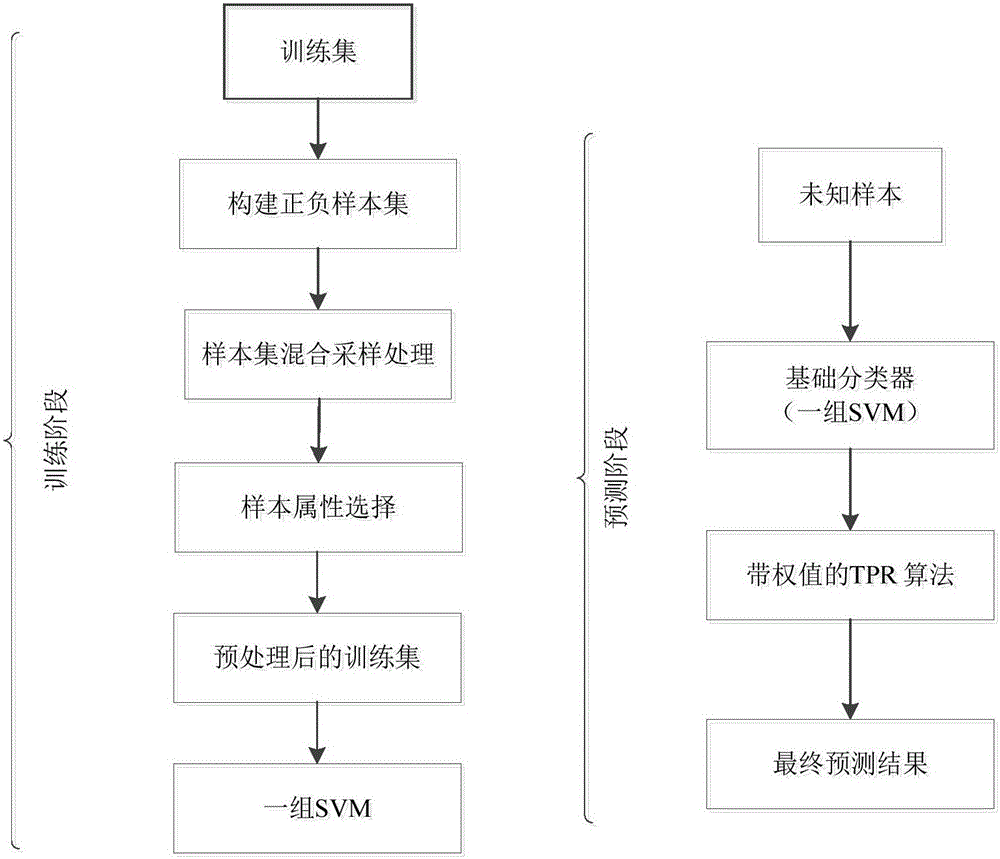
The invention relates to the field of bioinformatics and data mining, in particular to a hierarchical multi-label classification method for protein function prediction, and aims at solving the data set imbalance problem, multi-label problem and hierarchical constraint problem when the conventional classification methods are used for predicting protein functions. The method comprises the following steps of: 1, a training stage: training a data set of each node in a class label hierarchical structure by adopting an SVM classifier in the training stage so as to obtain a group of basic classifiers; and 2, a prediction stage: firstly obtaining preliminary results of unknown samples by using the group of basic classifiers obtained in the training stage in the prediction stage, and processing the results by adopting a TPR algorithm with a weight so as to obtain a final result which satisfies a hierarchical constraint condition and realize the prediction of the protein functions. The hierarchical multi-label classification method for protein function prediction is applied to the field of bioinformatics and data mining.
Owner:NAT INST OF ADVANCED MEDICAL DEVICES SHENZHEN
Method for screening compound with targeted action on inactive conformation of protein kinase
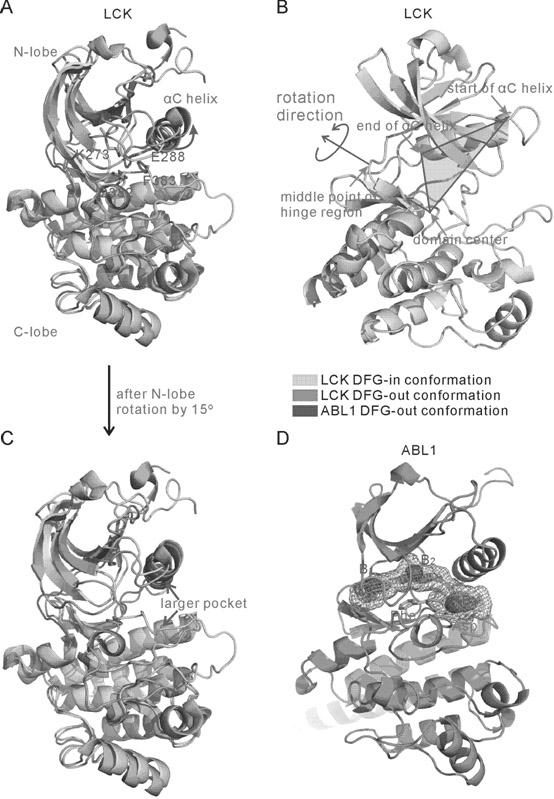
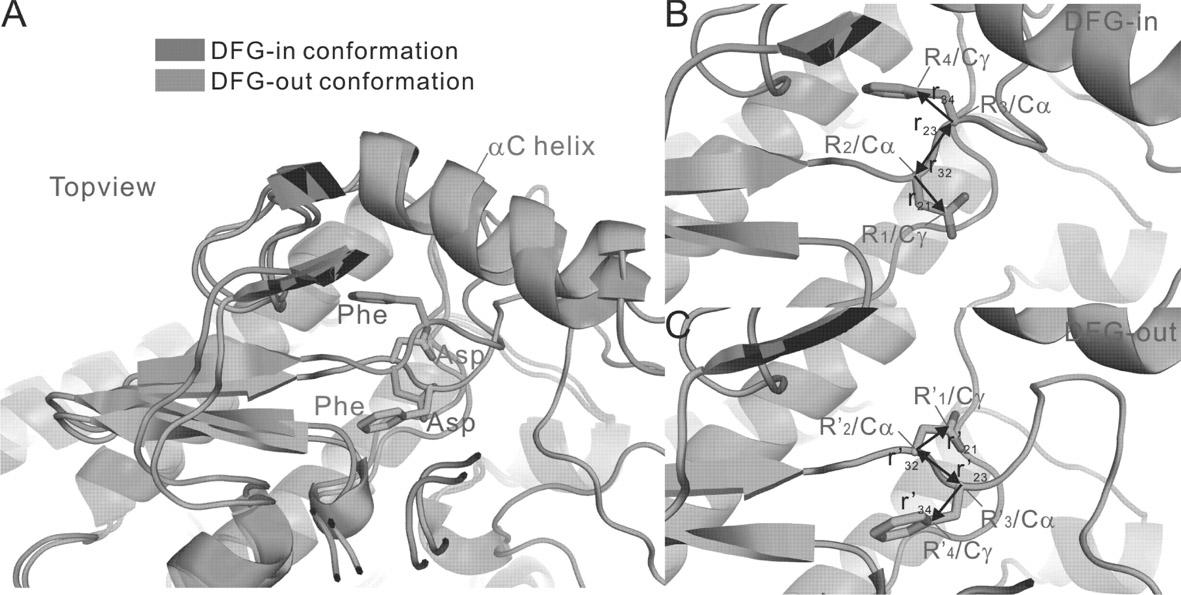
The invention belongs to the technical field of protein structure prediction and drug molecule virtual screening, specifically to a method for screening a compound with targeted action on an inactive conformation of a protein kinase. The method provided by the invention comprises a prediction method for the conformation of an active chain section of the protein kinase, wherein a corresponding DFG-out inactive conformation is generated from the DFG-in active conformation of the protein kinase; the method also comprises a selection method of a combined conformation after implementing butt joint of a II type inhibitor, and the selection method is used for selecting small molecules in conformation prediction and virtual screening. The method for screening a compound with targeted action on an inactive conformation of a protein kinase is already calculated and verified in the protein kinases of seven types of known inactive conformations, wherein the success rate is close to 96%. The method provided by the invention is already applied to the prediction of inactive conformation of PknB protein kinase of tubercle bacillus and the virtual screening of a possible II type inhibitor of PknB, and the bacteriostasis of two kinds of small molecules are already found according to bacteriostatic experiments.
Owner:FUDAN UNIV
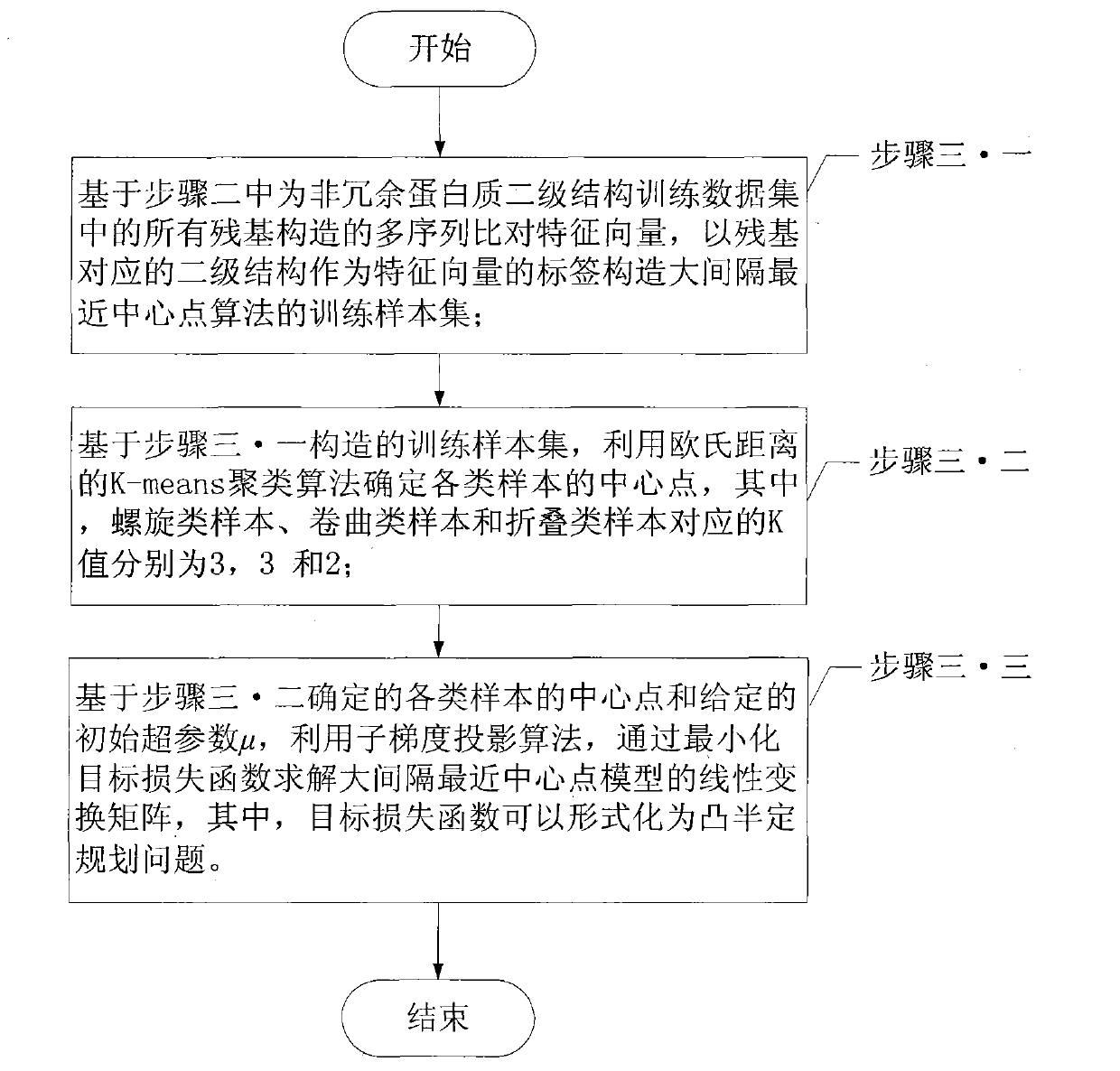
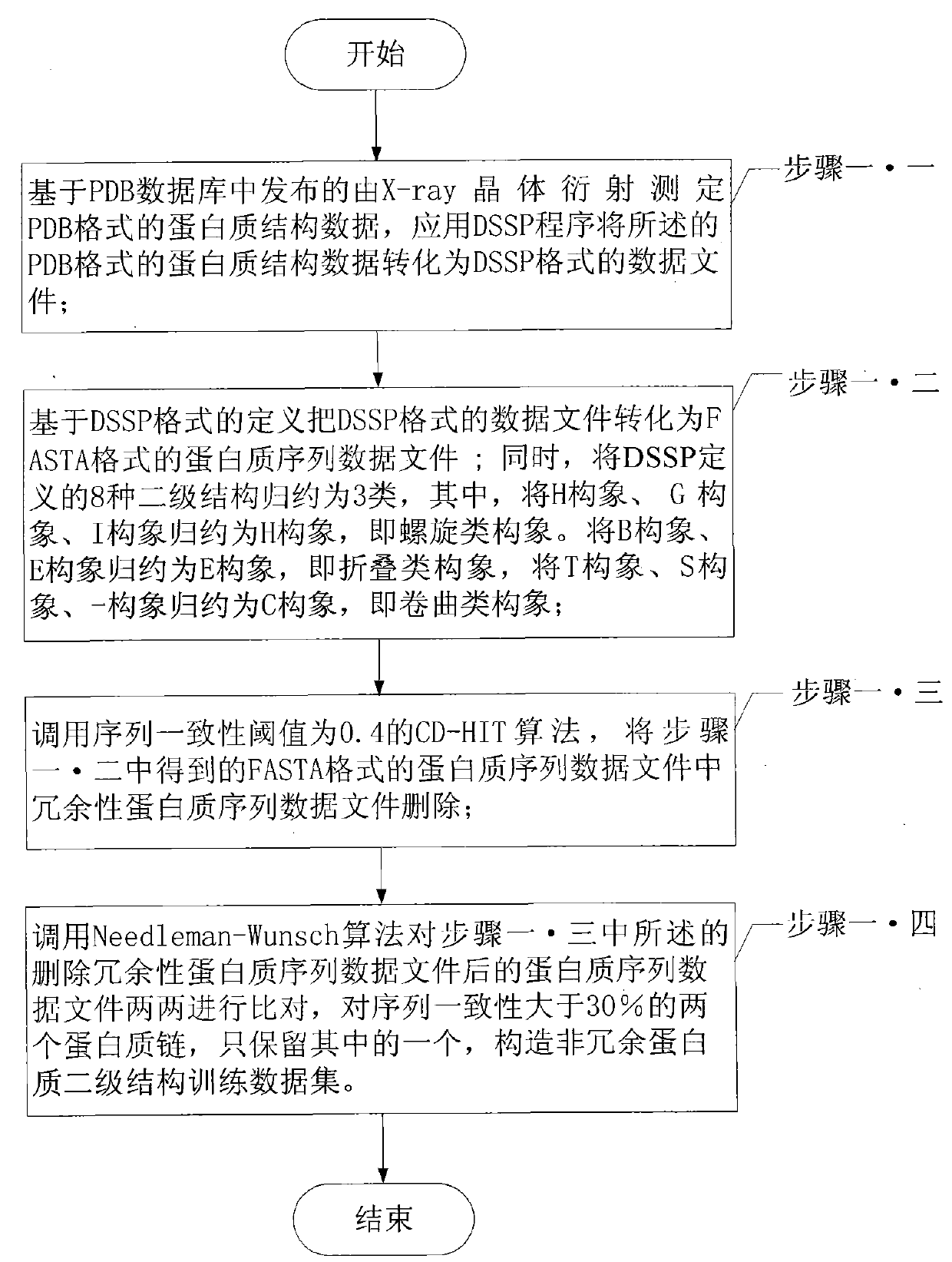
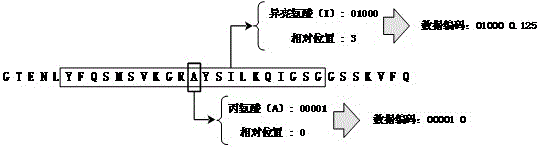
Protein secondary structure engineering prediction method based on large margin nearest central point
The invention relates to a protein secondary structure engineering prediction method based on large margin nearest central point, belonging to the protein secondary structure engineering prediction method field and solving the problems that the existing protein secondary structure prediction method has local minimum of data weight number and low prediction efficiency when adopting machine learning algorithm. The protein secondary prediction method of the invention includes that: firstly a non-redundant protein secondary structure training data set is constructed based on PDB database, then a multi-sequence comparison characteristic is constructed for a target protein chain based on NCBT nr database, and finally the large margin nearest central point algorithm is utilized to build a protein secondary structure prediction model. The large margin nearest central point algorithm utilizes Euclidean distance K-means clustering algorithm to determine the central point of each sample, and linear transformation of input space is learned by a minimization target loss function. The invention realizes fast, high-efficiency and high-precision protein secondary structure prediction and is applicable to protein secondary structure prediction.
Owner:HARBIN INST OF TECH
BP neural network based protein secondary structure prediction method
InactiveCN105740646AFast convergenceIncrease parallel spaceSpecial data processing applicationsNeural learning methodsAdaptive learningNetwork output
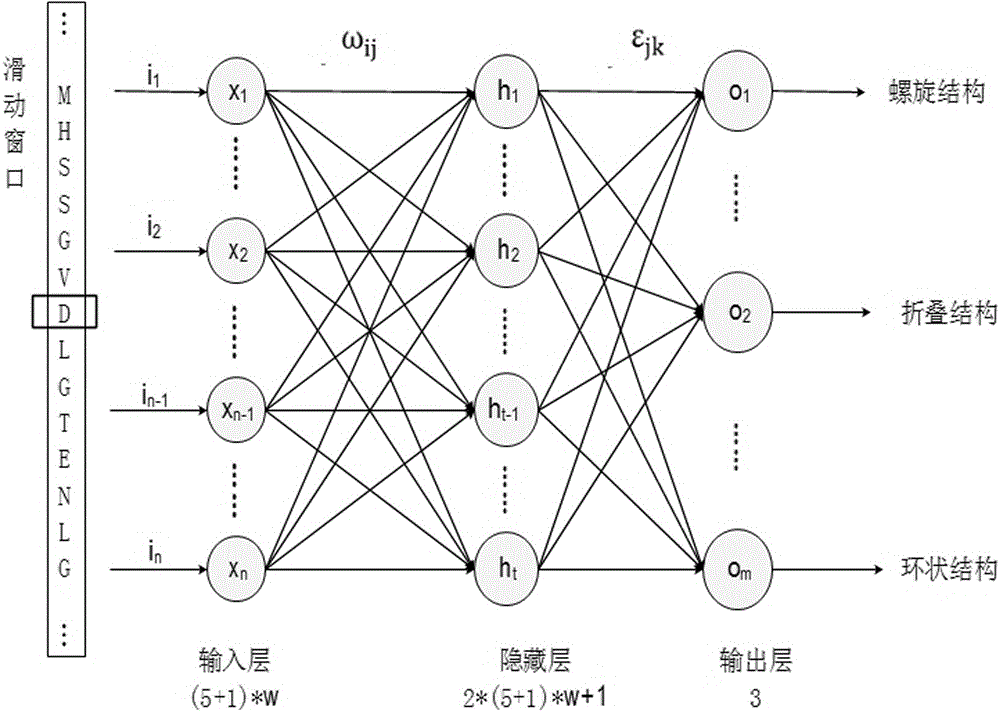
The invention belongs to the field of protein secondary structure prediction methods, relates to a BP neural network training and prediction method used for protein secondary structure prediction and solves the problem of bad prediction effect of the protein secondary structure. The BP neural network training and prediction method comprises the steps of firstly selecting a group of training sample sets with [alpha]-helix, [beta]-sheet and coiling structures accounting for normal proportions from PDB, coding an amino acid sequence of a protein and regarding the coded amino acid sequence of the protein as a network input, and regarding a secondary structure of the corresponding amino acid as a network output; optimizing based on a gradient method, introducing a learning rule which is attached with a momentum item and a self-adaptive learning rate to avoid an oscillation phenomenon and prevent from being trapped in a local minimum value; adopting a six-bit input coding way and a sliding window technology in an input layer, setting a hidden layer structure based on an experience formula and the size of a sliding window; and outputting and predicting classification of the protein secondary structure by an output layer based on a DSSP algorithm.
Owner:HUNAN UNIV OF TECH +1
Protein structure prediction method based on tree structure replica exchange and fragment assembly
ActiveCN104200130ALower search costImprove efficiencySpecial data processing applicationsTree (data structure)Native state
The invention discloses a protein structure prediction method based on tree structure replica exchange and fragment assembly. The method includes the following steps that protein conformations are based on a replica exchange thought, on the basis that preservation of a plurality of replica messages is facilitated, protein conformation space search can be enhanced, the advantage of a tree data structure is used, energy layering and protein middle conformation shape information can also be used, a specific weight function is used for selecting the entire protein confirmation spaces through certain probability as compiled replicas, the protein conformations are led to step towards a low-energy path, the diversity of the protein conformations can be reserved to the greatest extent, and thus the purpose for predicting the protein native state can be achieved ultimately. By means of the protein structure prediction method based on tree structure replica exchange and fragment assembly, the calculated amount can be greatly reduced, computing time is shortened, and meanwhile it is guaranteed that the low-energy conformations can be searched for.
Owner:ZHEJIANG UNIV OF TECH
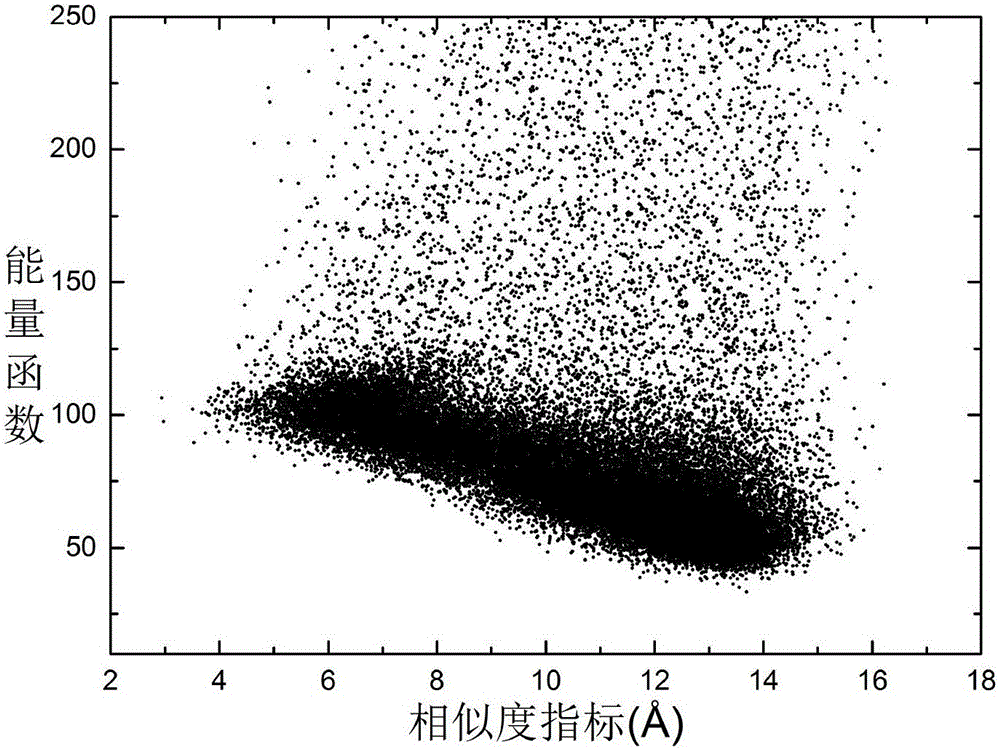
Local Lipschitz support surface-based dual-layer differential evolution protein structure prediction method
ActiveCN106096328AFast convergenceImprove reliabilitySpecial data processing applicationsMolecular structuresComputer scienceSupport surface
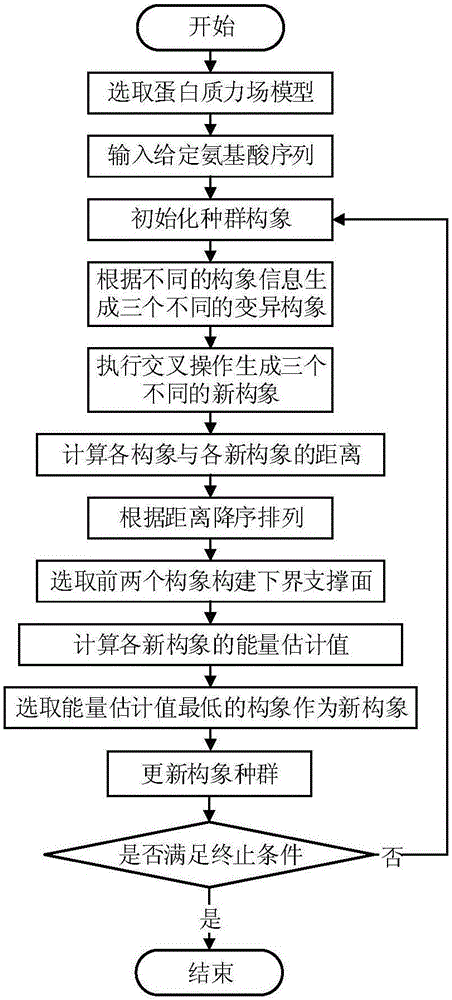
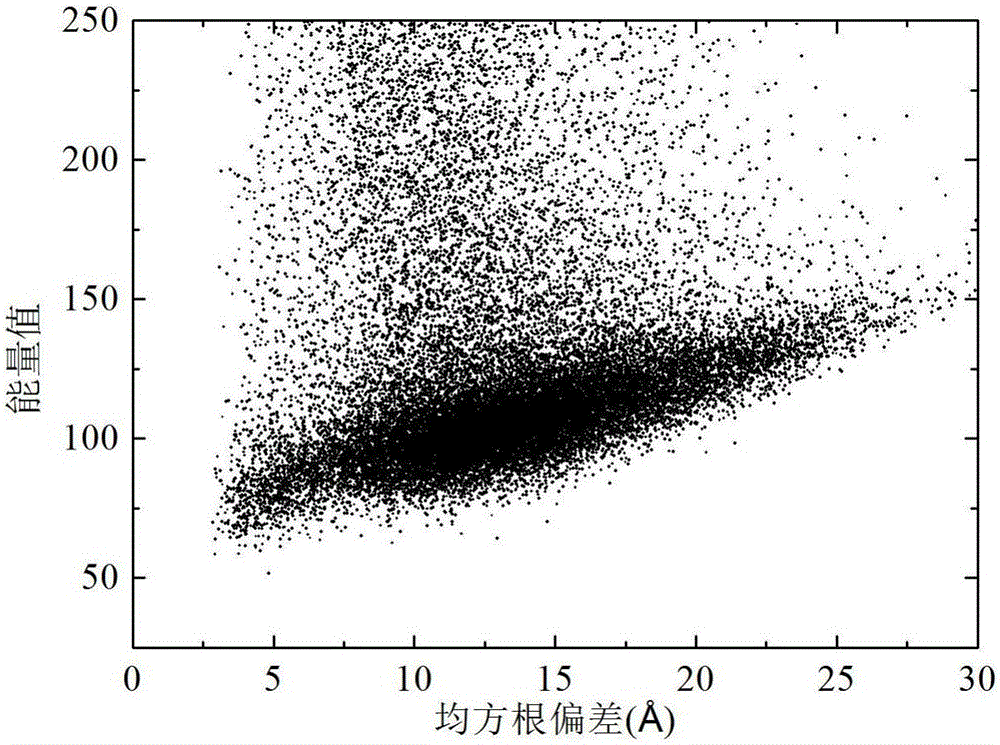
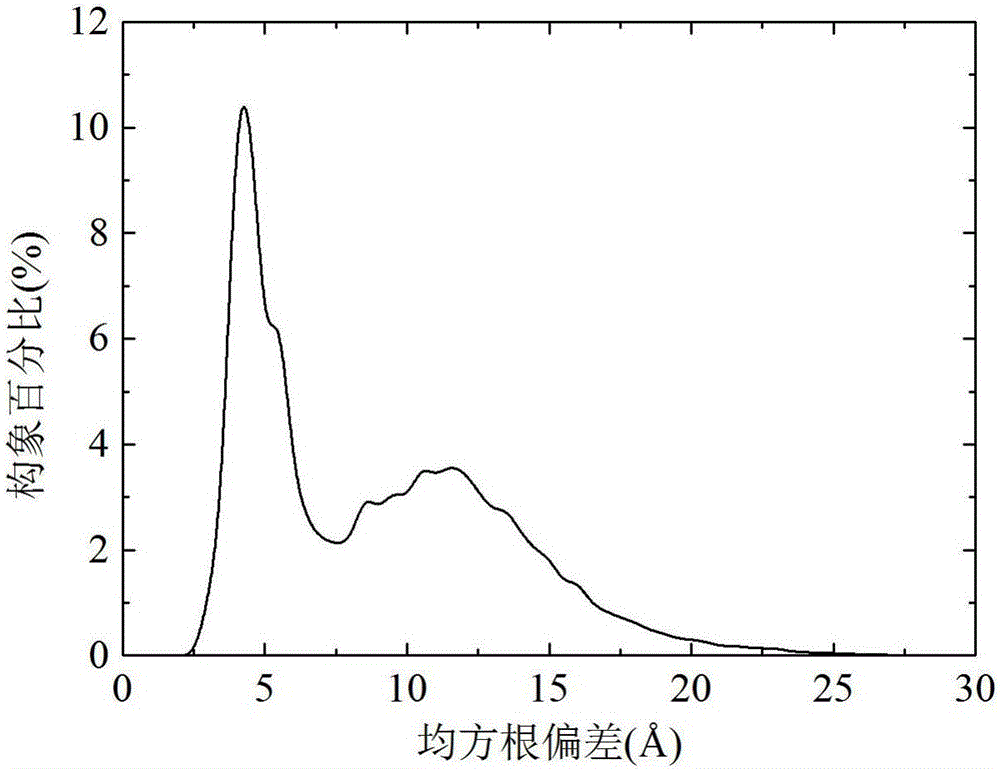
The invention discloses a local Lipschitz support surface-based dual-layer differential evolution protein structure prediction method. The method comprises the steps of firstly, selecting an optimal conformation in a current population according to an energy value, calculating distances from other conformations to the optimal conformation, and ranking all the conformations according to the distances; secondly, selecting part of the conformations closest to the optimal conformation to establish a Lipschitz lower bound support surface, calculating an energy lower bound estimation value of each selected conformation, and calculating an average error of an actual energy value and the lower bound estimation values; and finally, dividing an algorithm into two layers according to the average error, randomly selecting the conformation to perform fragment assembling to generate a new conformation by the first layer, and performing fragment assembling according to the optimal conformation to generate a new conformation by the second layer, so as to guide the algorithm to be quickly and reliably converged to a region with the lowest energy. The method is high in prediction precision and relatively low in calculation cost.
Owner:ZHEJIANG UNIV OF TECH
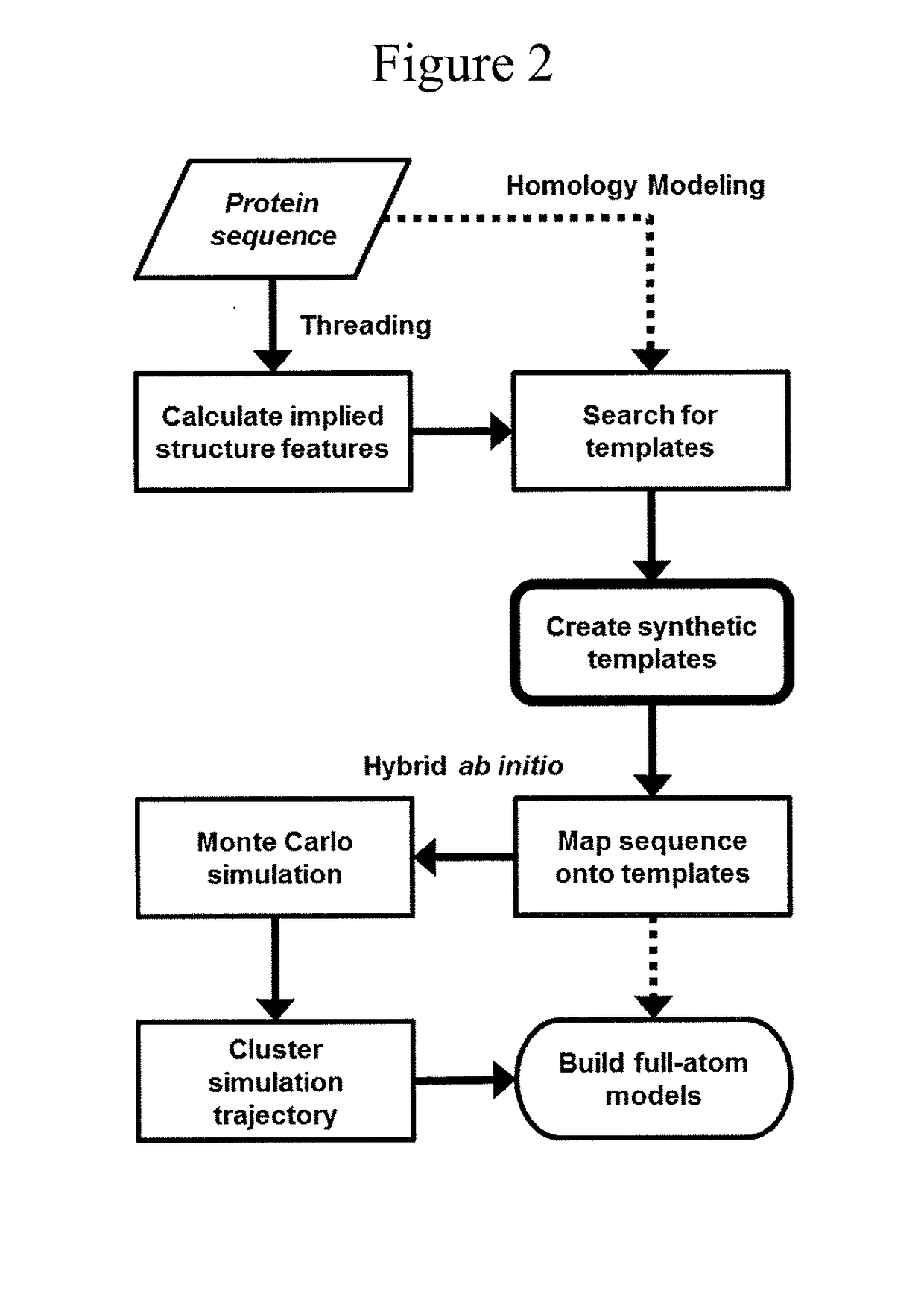
Protein structure prediction system
ActiveUS20180260517A1Increase structural diversityImprove discovery rateChaos modelsNon-linear system modelsEnergy minimizationTarget peptide
The present invention is an accelerated conformational sampling method for predicting target peptide and protein structures comprising a process of determining energy minimized synthetic templates using a simple system for modeling individual molecular bonds within the subject peptide or protein. Use of these synthetic templates greatly reduces the computational resources necessary for optimally determining structural features of the target peptide or protein. The present invention also provides methods for rapid and efficient analysis of the effect of mutations on target peptides and proteins.
Owner:DNASTAR
Method for predicting protein structure on basis of two-stage differential evolution algorithm
InactiveCN105760710AImprove forecast accuracyReduce complexitySpecial data processing applicationsMolecular structuresData miningPhases of clinical research
The invention discloses a method for predicting the protein structure on the basis of a two-stage differential evolution algorithm. The method comprises the following steps: under a framework of the differential evolution algorithm (DE), firstly carrying out random folding and disturbance on an inputted inquiry sequence, and generating initial conformation populations with diversified folding types; then dividing conformation searching into two stages according to iterative times; in the first stage, randomly selecting one conformation from the populations as a target individual; in the second stage, dividing the population into two parts according to energy, and randomly selecting an individual from the front 50% of populations with low energy as a target individual; then randomly selecting three conformation individuals different from the target individual, and generating a testing individual by variation, crossing and a segment assembling strategy; when the populations are updated, judging whether the testing individual is accepted according to the energy of the conformation; and under the guidance of the two staged population, obtaining a series of metastable-state conformations with higher predicting accuracy and lower complexity by continuously updating the populations. The method disclosed by the invention has the advantages of higher predicting accuracy and lower complexity.
Owner:ZHEJIANG UNIV OF TECH
Inherent irregular protein structure forecasting method based on kernel canonical correlation analysis
InactiveCN102779240AGood forecastImprove forecast accuracySpecial data processing applicationsPharmacyKernel canonical correlation analysis
The invention provides an inherent irregular protein structure forecasting method based on kernel canonical correlation analysis. The method includes: (1) extracting architectural features and biochemical features of protein to be forecasted to serve as recognition features, wherein the architectural features are combination frequencies of amino acid on the periphery of forecasting sites of protein and obtained in a window method, and the biochemical features are Russell / Linding value, hydrophobicity, polarity and electrification of amino acid at the forecasting sites of protein; (2) performing mapping and integrating for the extracted feature data in the kernel canonical correlation analysis method to obtain feature data favorable for protein structure recognition, wherein a kernel function used in the kernel canonical correlation analysis method is a radial basis function; and (3) recognizing and forecasting protein structure on the basis of the feature data favorable for protein structure recognition. The feature data favorable for protein structure recognition is effectively improved in forecasting accuracy, is favorable for supplying early-period basis to finding and verification of inherent irregular protein, and supplies foundation to development of biological pharmacy.
Owner:HARBIN ENG UNIV
Method for predicting protein structure from local to global on basis of knowledge spectrum
InactiveCN105808972AGood conformational space sampling abilityImprove forecast accuracySpecial data processing applicationsMolecular structuresComputer scienceProtein structure prediction
The invention discloses a method for predicting a protein structure from local to global on the basis of a knowledge spectrum. The method comprises the following steps: for a query sequence, obtaining a high-quality fragment library through a multi-feature seamless threading method, and obtaining distance spectrum knowledge between residues through statistic consistency analysis on the basis of the fragment library; dividing the query sequence into several segments of structures according to residue information recorded in a distance spectrum; carrying out fragment assembling on each segment of structure to obtain relatively low energy and to ensure that the space distance between adjacent residues approaches to a predicted distance in the distance spectrum; carrying out fragment assembling on the unsegmented structures and calculating the global energy to obtain a metastable-state conformation with low energy and more reasonable structure. The method disclosed in the invention is relatively good in conformation space sampling ability and high in prediction precision.
Owner:ZHEJIANG UNIV OF TECH
Protein structure prediction method and system based on deep learning
ActiveCN112233723AQuick buildBuild accuratelyBiostatisticsSequence analysisAlgorithmNetwork construction
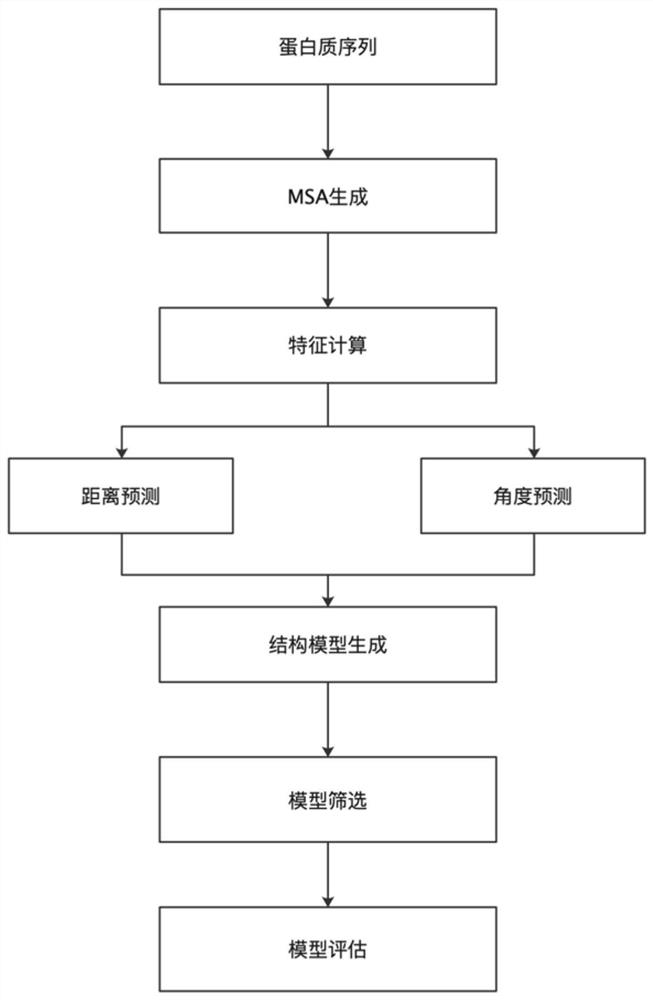
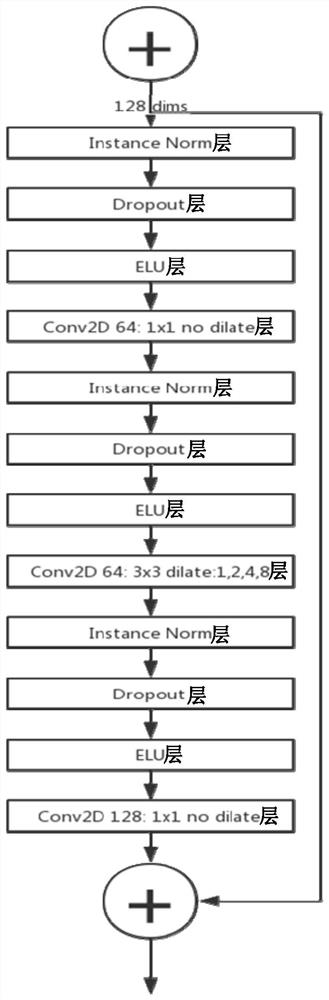
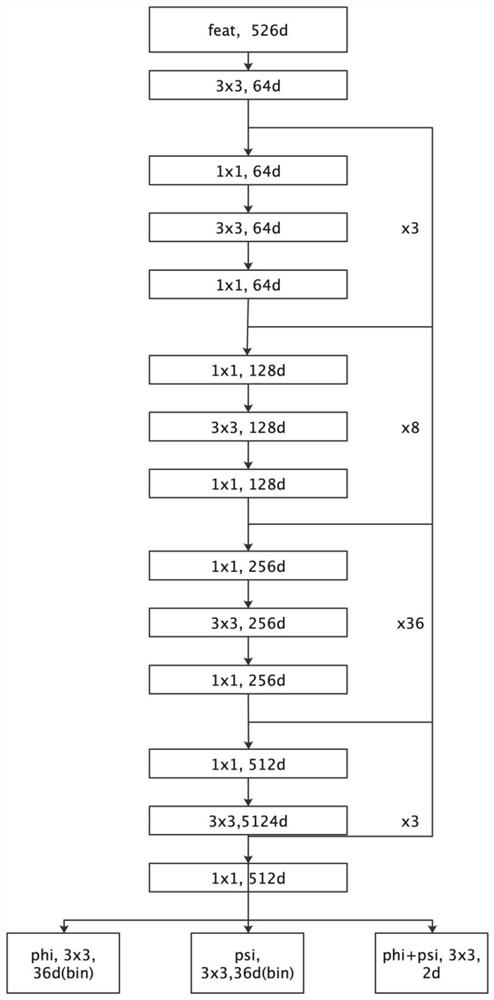
The invention provides a protein structure prediction method and system based on deep learning, and particularly relates to a protein three-dimensional structure simulation method based on deep learning and a biological information theory. The method comprises a protein homologous matrix search step, a related characteristic data calculation step, a protein residue distance and angle prediction network construction step, a distance and angle prediction accuracy evaluation step, a distance and angle-based three-dimensional model rapid generation and optimization step, a three-dimensional structure model screening step, and a prediction result evaluation step. Compared with a traditional method, the process has the advantages of being accurate and rapid in prediction, and high-throughput macroproteome simulation can be carried out.
Owner:上海天壤智能科技有限公司 +1
Deep learning Residue2vec-based protein structure prediction method
ActiveCN106372456AImprove matchImprove forecast accuracyNeural architecturesSpecial data processing applicationsAlgorithmTheoretical computer science
The invention discloses a deep learning Residue2vec-based protein structure prediction method. The method comprises the following steps of: giving input sequence information, regarding a known protein structure on a PDB website as a corpus to train, partitioning the proteins with known structures into residues with lengths of n, obtaining the expression of each residue in a vector space through a CBOW model and a Huffman code, and judging the similarities between the residues through calculating the distances between residue vectors, so as to obtain the front N fragment structures on each residue position of a query sequence and then form a fragment library of Residue2vec; carrying out random folding on the query sequence to form an initial conformation; randomly selecting a residue with the length of n, and carrying out dihedral angle replacement on the residue and fragments in the fragment library; and comparing the energy, if the energy is decreased, receiving the conformation, and if the energy is increased, receiving the conformation via a Metropolis criterion and finally obtaining a metastable-state conformation through continuous iteration. According to the method disclosed by the invention, the matching degree and prediction precision in the query sequence are relatively high.
Owner:ZHEJIANG UNIV OF TECH
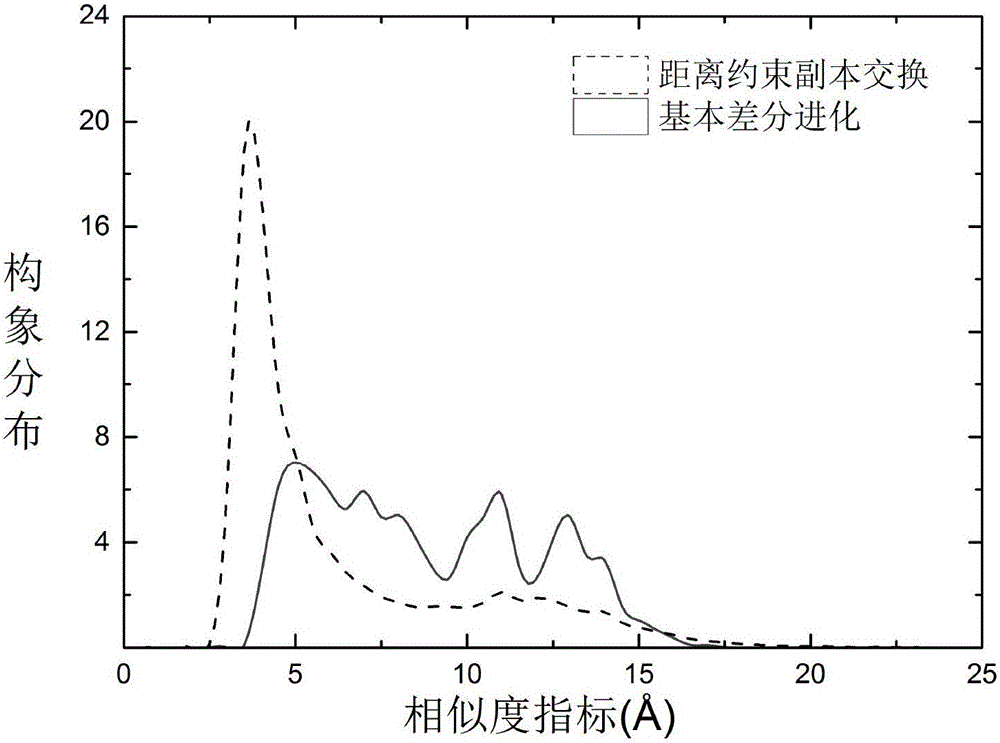
Method for predicting protein structure based on phased multi-strategy copy exchange
ActiveCN106055920AImprove forecast accuracyReduce complexitySpecial data processing applicationsMolecular structuresMetapopulationPhases of clinical research
This invention discloses a method for predicting protein structure based on phased multi-strategy copy exchange. In the frame of differential evolution (DE), generate initial conformation populations with a variety of folding types in each temperature layer; to each temperature layer, divide a conformation search into two phases according to iterations; randomly select a conformation as a target unity from the populations in the first phase; divide the populations into two parts according to energy in the second phase; randomly select one unity as the target unity from first 50% of populations with low energy; randomly select three conformation unities different from the target unity to generate a testing unity through variation, cross and section combination strategy; judge whether to accept the testing unity according to the energy of the conformation so as to perform copy exchange to corresponding unities of adjacent temperature layers; and, under the guidance of the phased strategy and the supplement of copy exchange strategy, acquire a series of metastable conformation through updating continuously.
Owner:ZHEJIANG UNIV OF TECH
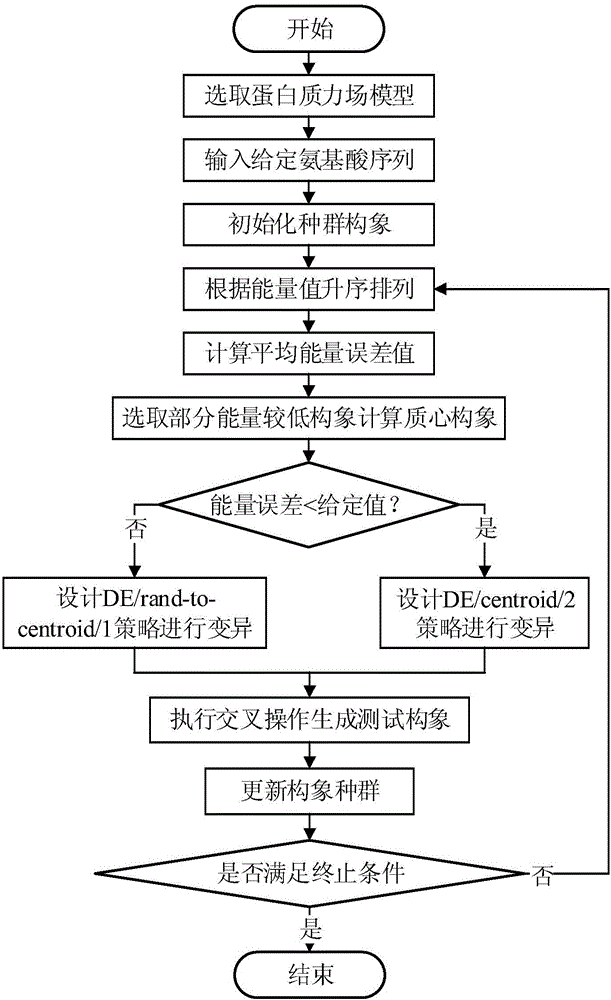
Centroid mutation strategy-based differential evolution protein structure prediction method
ActiveCN106096326AImprove search efficiencyImprove forecast accuracySystems biologySpecial data processing applicationsProtein structure predictionCentroid
The invention discloses a centroid mutation strategy-based differential evolution protein structure prediction method. The method comprises the steps of firstly, performing ascending sort according to an energy value of each conformation and calculating an average energy error value of each conformation and the conformation with the lowest energy; secondly, selecting part of the conformations with the relatively low energy to calculate a centroid conformation; and finally, judging a search state of an algorithm according to the average energy error value so as to design different centroid mutation strategies for generating test conformations, namely, if the average energy error value is greater than a set threshold, designing a DE / rand-to-centroid / 1 strategy to perform mutation, replacing corresponding segments in the randomly selected conformation by extracting partial segments in the centroid conformation to generate the test conformation, or otherwise, designing a DE / centroid / 2 strategy to perform mutation, replacing the corresponding segments in the centroid conformation by extracting the segments in the randomly selected conformation to generate the test conformation. Therefore, the search efficiency and prediction precision of the algorithm are improved.
Owner:ZHEJIANG UNIV OF TECH
Protein structure prediction method based on distance constraint copy exchange
InactiveCN105975806AIncrease diversityImprove forecast accuracySpecial data processing applicationsMolecular structuresAlgorithmEnergy analysis
The invention discloses a protein structure prediction method based on distance constraint copy exchange. The protein structure prediction method comprises the following steps: firstly, carrying out random folding and transformation on a query sequence in each temperature layer to generate an initial population; in population update, taking Rosetta Score3 as an optimal object function, and taking each individual in the population as a target individual in each temperature layer on the basis of a structure with lowest free energy when a protein native state structure which is put forward by Anfinsen is adopted; then, randomly selecting two individuals different from the target individual to carry out variation and cross to generate variation individuals, randomly selecting one section from another individual to carry out transformation with the variation individual to generate a test individual; carrying out energy value comparison on the test individual and the target individual, and introducing the knowledge of a distance spectrum for the test individual of which the energy rises; and carrying out copy exchange on the corresponding individual of an adjacent temperature layer. The protein structure prediction method has good conformational space sampling capability and high prediction accuracy.
Owner:ZHEJIANG UNIV OF TECH
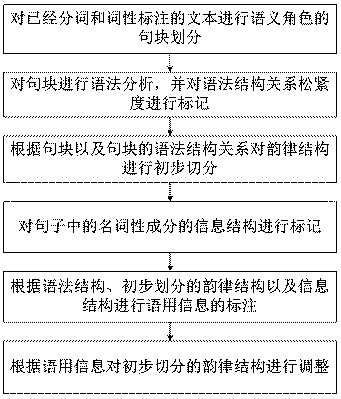
Chinese rhythm structure prediction method for combining with syntax semantic pragmatic information
ActiveCN108470024AFully descriptiveImprove predictive performanceSemantic analysisSpeech recognitionFeature extractionSpeech synthesis
The invention belongs to the field of speech synthesis, and particularly relates to a Chinese rhythm structure prediction method for combining with syntax semantic pragmatic information. The method aims to combine the syntax semantic pragmatic information with a rhythm structure prediction model through the analysis of multidimensional features which affect the rhythm structure and a relevant action mode thereof so as to improve the nature degree of the speech synthesis. The method comprises the following steps that: according to semantic role tagging, carrying out sentence block division; carrying out grammatical analysis on the sentence block, and tagging the degree of tightness of the relationship of a syntactic structure; carrying out preliminary segmentation on the rhythm structure; tagging the information structure of a noun composition in the sentence; tagging pragmatic information; and according to the pragmatic information, regulating the rhythm structure subjected to preliminary segmentation. By use of the method, deep syntax and semantic role tagging and the pragmatic information are introduced into the text tagging system, text tagging, feature extraction and rhythm structure prediction are realized through a way of combining statistics with rules, and the accuracy of a rhythm prediction model is effectively improved.
Owner:北京灵伴即时智能科技有限公司
Multi-strategy colony protein structure prediction method based on local abstract convexity supporting plane
ActiveCN106650305AReduce the averageAvoid premature convergenceSequence analysisSpecial data processing applicationsAlgorithmTheoretical computer science
The invention discloses a multi-strategy colony protein structure prediction method based on a local abstract convexity supporting plane. The method comprises the steps of firstly generating three different new conformation individuals according to randomly chosen conformation individuals, a current target conformation individual and a conformation individual with a minimum energy value; then calculating the distance of each conformation individual in a current colony to each new conformation individual, and conducting ascending sort according to the distance; next, calculating an abstract convexity supporting plane of a partial conformation individual which is closest to each new conformation individual, and thus calculating an energy estimated value of each new conformation individual; finally, comparing the energy estimated value of each new conformation individual and thus choosing a new conformation individual with minimum energy estimated value to conduct energy function evaluation. The provided multi-strategy colony protein structure prediction method based on the local abstract convexity supporting plane is high in prediction accuracy and high in search efficiency.
Owner:ZHEJIANG UNIV OF TECH
Protein structure prediction method based on improved niche genetic algorithm
InactiveCN105184112AIncrease diversityAvoid duplicate solutionsSpecial data processing applicationsMinimum free energyAlgorithm
The invention relates to the field of protein structure prediction and discloses a protein structure prediction method based on the improved niche genetic algorithm. According to the method, the niche genetic algorithm is introduced into protein structure prediction, and the genetic algorithm is improved to a certain extent in terms of selection and variation. Based on data obtained from experiments and results obtained through comparison with other methods, the method has the advantages that the corresponding minimum free energy value of protein can be searched more comprehensively, so that a more stable protein structure is obtained; operation time is shortened greatly, meaning that the method has high time efficiency.
Owner:DALIAN UNIVERSITY
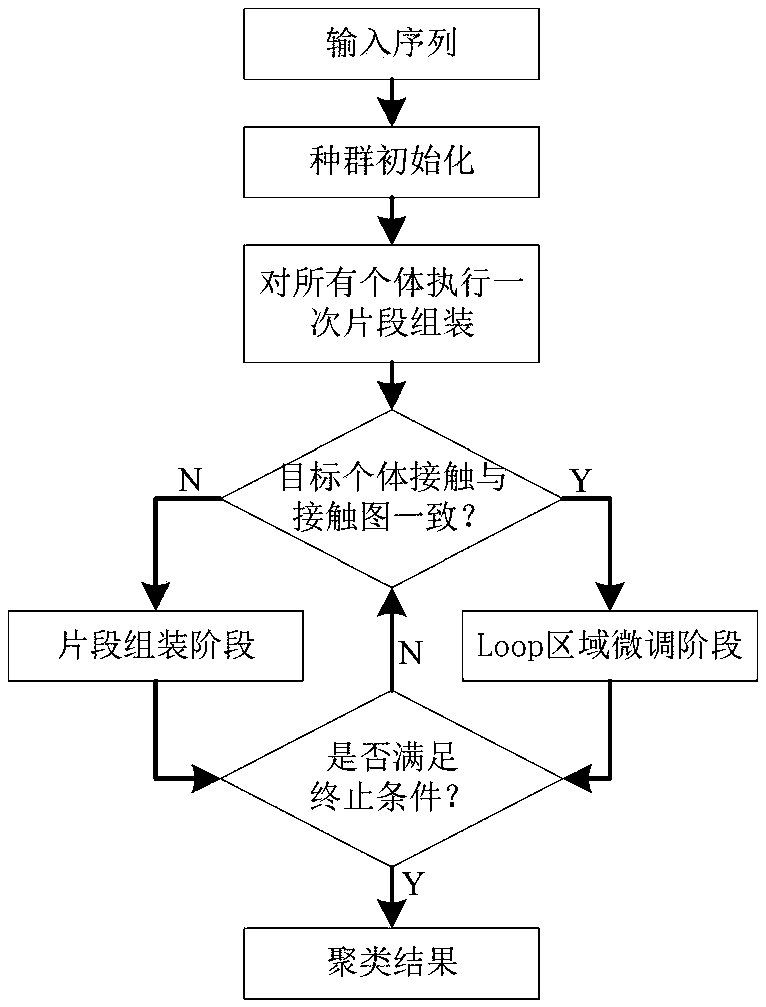
Protein structure prediction method based on residue contact information crossover strategy
ActiveCN109360599AImprove search efficiencyImprove forecast accuracyInstrumentsMolecular structuresSpecific testData mining
A protein structure prediction method based on a residue contact information crossover strategy is disclosed. The method comprises the following steps of firstly, using Robetta and RaptorX-Contact toacquire a fragment library and a contact map; secondly, using contact map information so that population individuals dynamically enter into a segment assembling phase and a loop area fine-tuning phase, and using the crossover strategy of a specific test individual and a specific area; and finally, through clustering, acquiring a final prediction result. Through using contact map information, population evolution is dynamically performed, and the crossover strategy of the specific area is used to increase search efficiency and improve prediction precision.
Owner:ZHEJIANG UNIV OF TECH
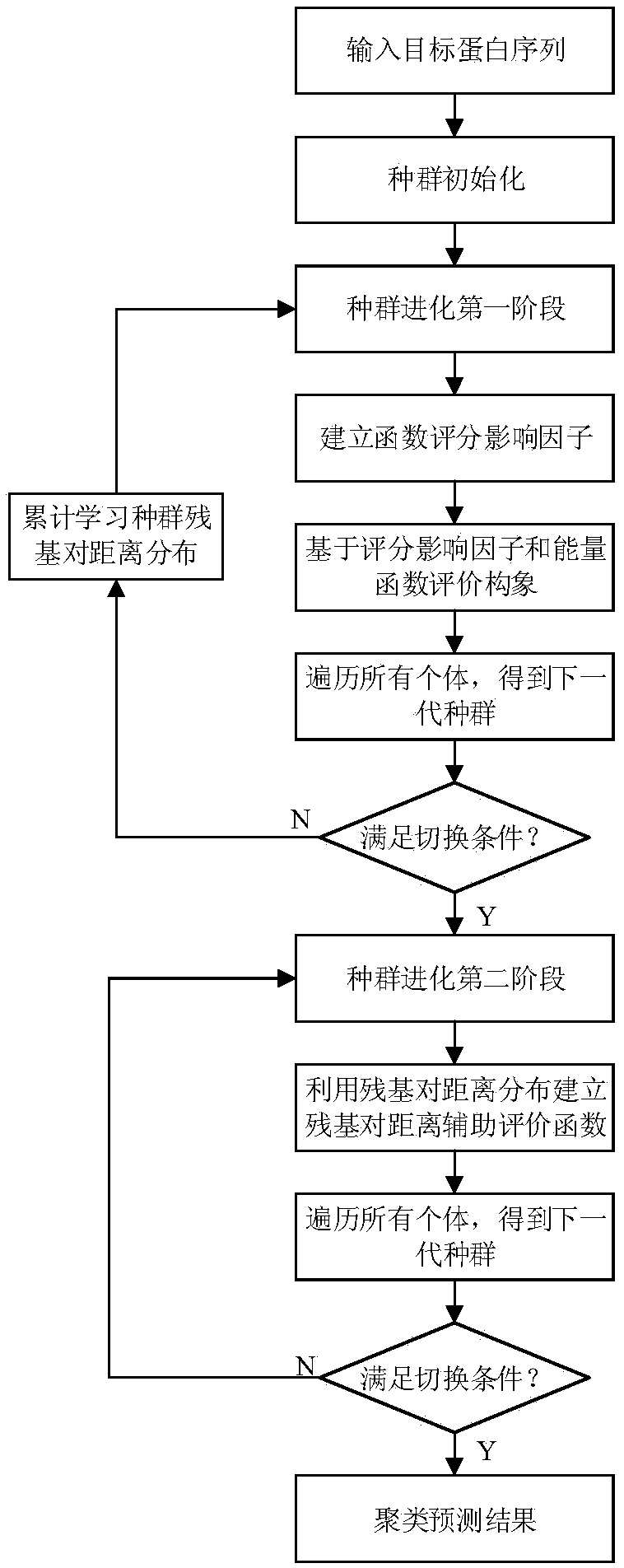
Residue contact information self-learning-based protein structure prediction method
ActiveCN109215732AEfficient conformational space searchTo achieve the purpose of dynamic switchingInstrumentsMolecular structuresEnergy functionalAutomatic learning
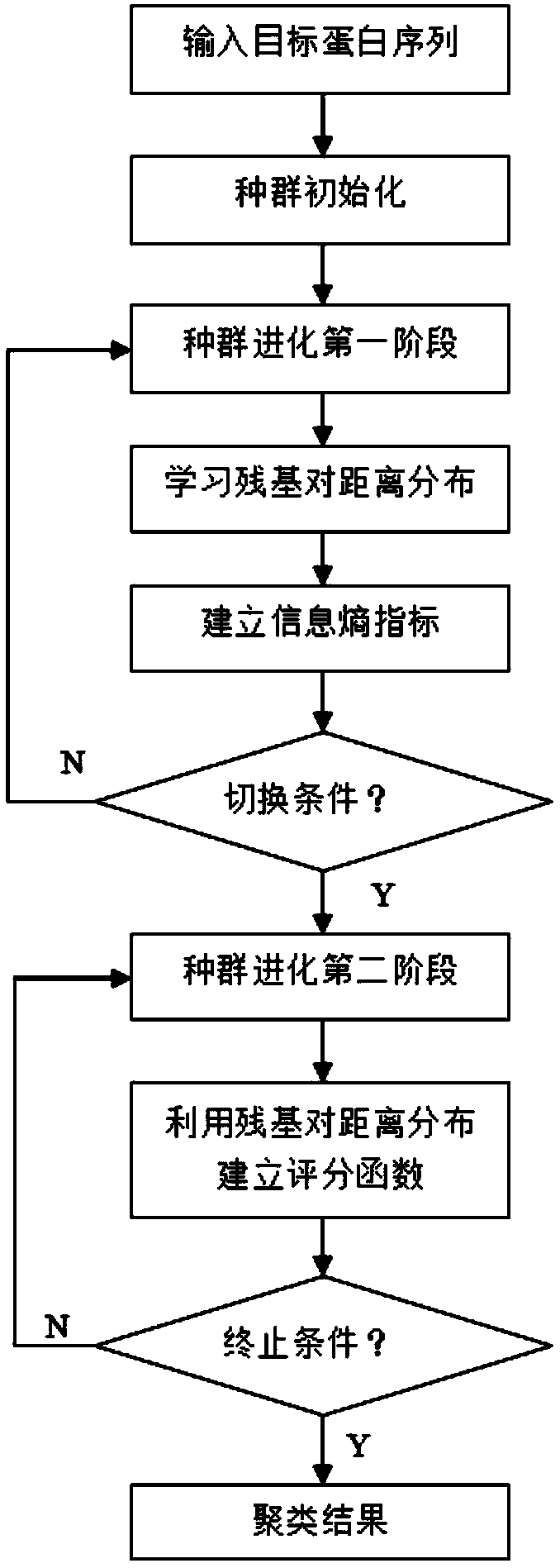
The invention relates to a residue contact information self-learning-based protein structure prediction method. According to the method, a fragment library and a contact map are obtained through Robetta and RaptorX-Contact; in the first stage of population evolution, residue pair distance distribution learning is performed, an information entropy index is established so as to reflect the convergence degree of a population, the purpose of automatic learning can be achieved; in the second stage of the population evolution, a scoring function is established with learned residue pair distance distribution information so as to assist an energy function in conformation space search; and a final prediction result is obtained through clustering. With the protein structure prediction method provided by the invention adopted, the residue pair distance information can be learned automatically so as to assist the energy function in conformation space optimization; and the information entropy indexis constructed, and therefore, the dynamic switching of the two stages can be realized.
Owner:ZHEJIANG UNIV OF TECH
Dihedral angle information auxiliary energy function selection based protein structure prediction method
ActiveCN109448784AReduce the impactEnhance local structureInstrumentsMolecular structuresNative structureEnergy functional
The invention discloses a dihedral angle information auxiliary energy function selection based protein structure prediction method. extracting dihedral angle information according to a Ramachandran plot corresponding to a protein residue; performing assessment on conformation by using the energy functions and the dihedral angle information in a Rosetta algorithm; giving different weights to two fractions, designing a new scoring function, and performing selection on the fragment assembled conformation by using the scoring function so that the influence on the three-dimensional structures of proteins due to inexact energy functions can be reduced; and performing global search and local search on the conformation, and performing enhancement on local structures on the basis of guaranteeing the global topological structure of the constellation so that more near-native structures can be obtained. Thus, the dihedral angle information auxiliary energy function selection based protein structure prediction method with good sampling capabilities and high prediction precision can be provided.
Owner:深圳新锐基因科技有限公司
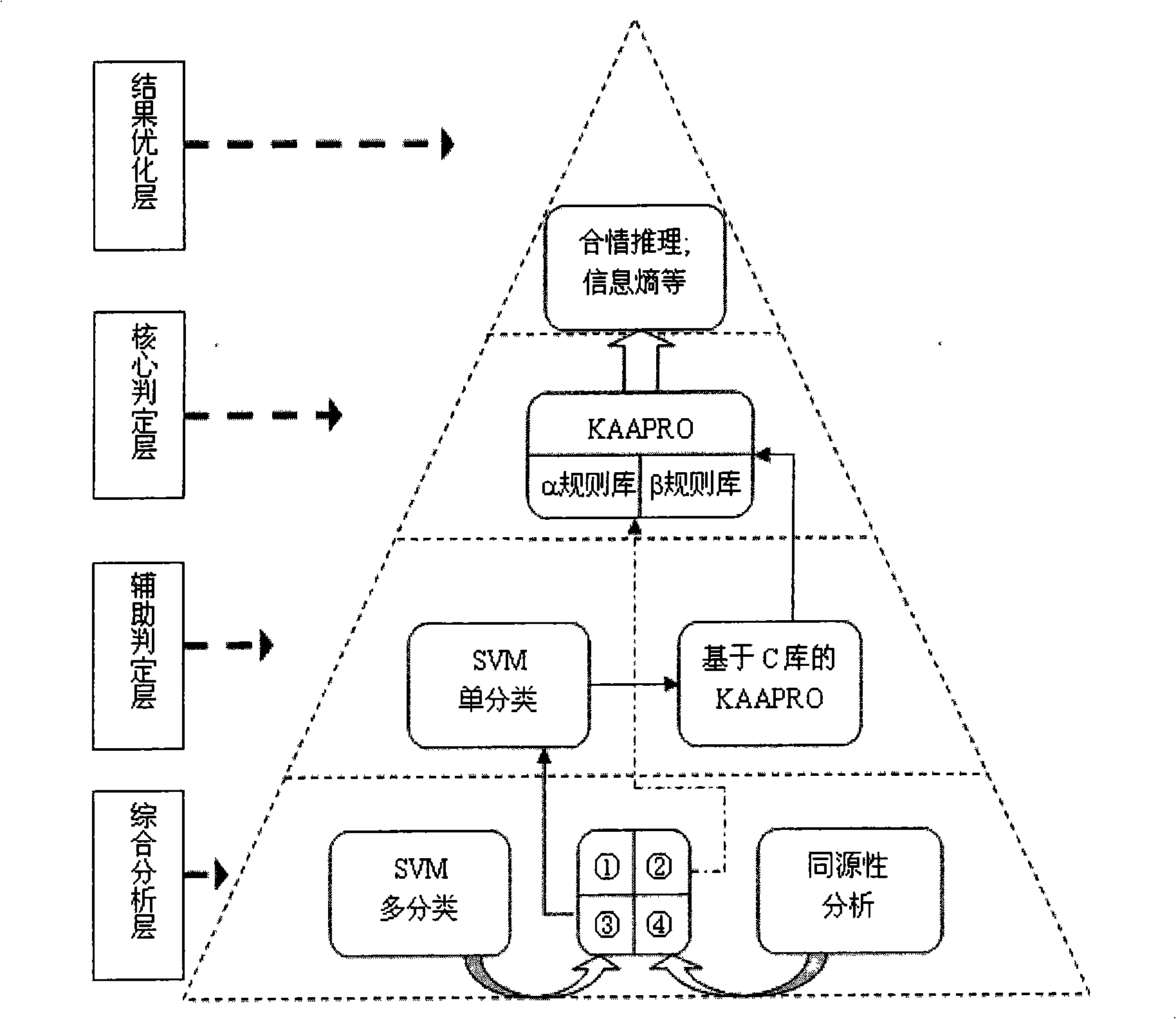
Secondary protein structure forecasting technique based on association analysis and association classification
InactiveCN101344902AImprove forecast accuracySignificant predictive performance advantageSpecial data processing applicationsClassification methodsCorrelation analysis
The invention discloses a protein secondary structure prediction technology based on correlation analysis and correlation classification, wherein, based on a double-base cooperating mechanism, a KDD process model is introduced into the problem of protein secondary structure prediction; in a KAAPRO method, data mining (knowledge discovery) is used as a main body and Maradbcm arithmetic based on the KDD process model and a D-CBA method of correlation rule classification are adopted. The correlation rule obtained by the KAAPRO method discloses the influence relation of amino acid physical-chemical properties on the protein secondary structure, thus enhancing the precision of prediction. The characteristic of the Maradbcm arithmetic on mining accident rules mines the correlation rules of alpha protein base and beta protein base which have relatively high purity, therefore, the obtained mining results are the distillated rules. The D-CBA correlation classification method uses the measure of credibility and supportability as a composite measure for carrying out the protein correlation classification. While guaranteeing the prediction precision, the technology provides a basis for the further analysis of the secondary structure for biologists.
Owner:UNIV OF SCI & TECH BEIJING
Residue contact information auxiliary evaluation-based protein structure prediction method
ActiveCN109215733ATopological optimizationImprove search efficiencyInstrumentsMolecular structuresContact mapPhases of clinical research
The invention relates to a residue contact information auxiliary evaluation-based protein structure prediction method. According to the method, a fragment library and a contact map are obtained through Robetta and RaptorX-Contact; the contact map is used to auxiliarily evaluate conformations with different ways in two stages of population evolution; and a final prediction result is obtained through clustering. According to the method of the invention, contact map information is adopted to auxiliarily evaluate conformations, and conformations with correct topology are reserved; and therefore, search efficiency and prediction accuracy can be improved. The residue contact information auxiliary evaluation-based protein structure prediction method of the invention has high prediction precision.
Owner:ZHEJIANG UNIV OF TECH
Population protein structure prediction method based on residue contact information
ActiveCN108846256AReduce forecast errorSpecial data processing applicationsProtein targetEnergy functional
A population protein structure prediction method based on residue contact information. Under the framework of the evolutionary algorithm, firstly, a test conformation is generated by exchanging fragments in the conformation. Secondly, the residue contact information of a target protein is predicted according the sequence information, and a residue contact energy function is designed to score the conformation; the residue contact energy is used to guide a selection process of the conformation, that is, if the energy of the test conformation is less than the energy of the target conformation, the test conformation is directly accepted, otherwise the residue contact energy is further compared; the residue contact energy of the test conformation is small, the test conformation is accepted, otherwise, acceptation is performed according to the Boltzmann probability so as to instruct the algorithm to sample a conformation with lower energy and a more reasonable structure. The conformation selection is guided by the residue contact energy, thereby alleviating the prediction error caused by the inaccuracy of the energy function. The invention provides a population protein structure prediction method based on residue contact information with high prediction accuracy.
Owner:ZHEJIANG UNIV OF TECH
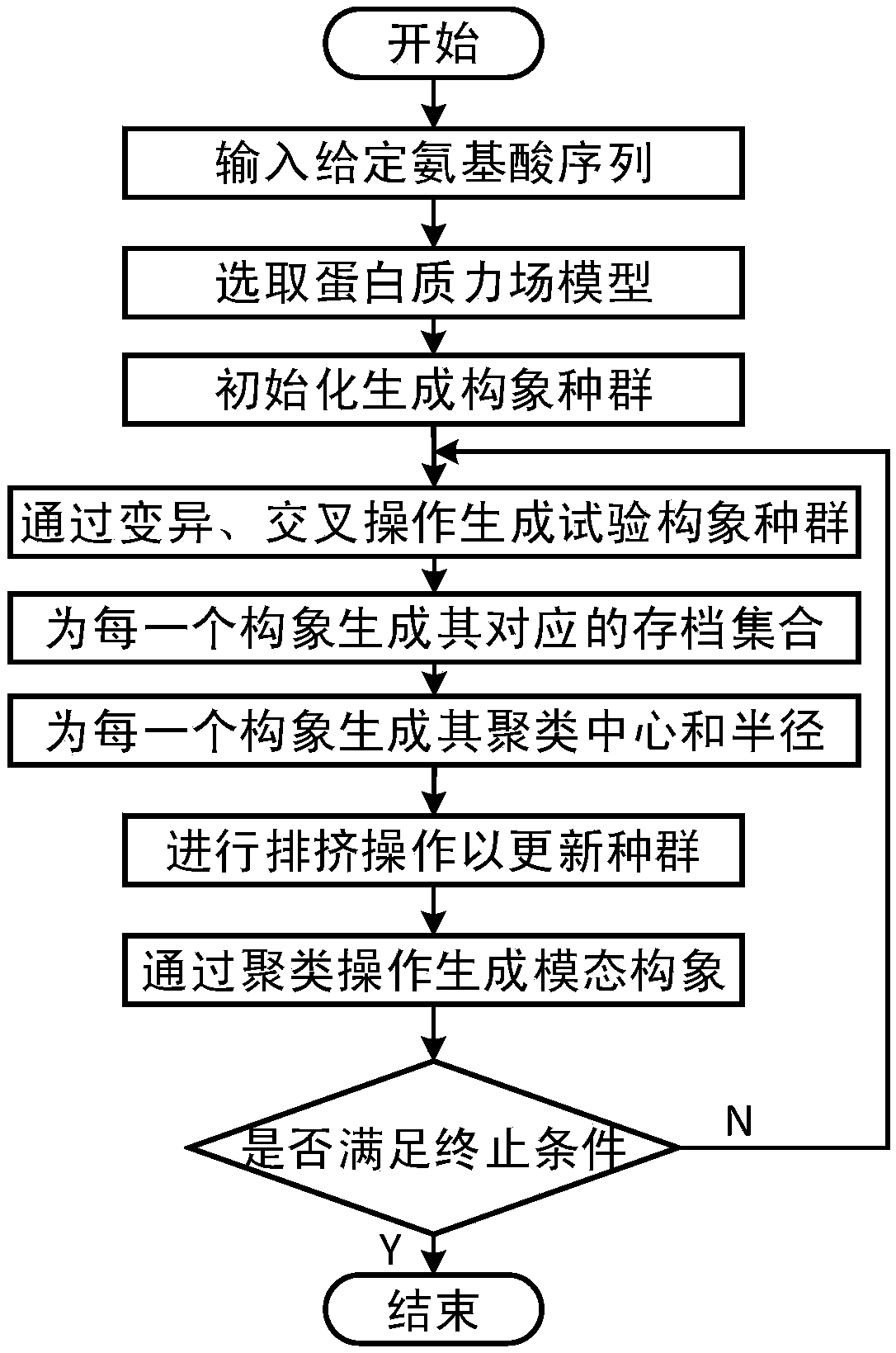
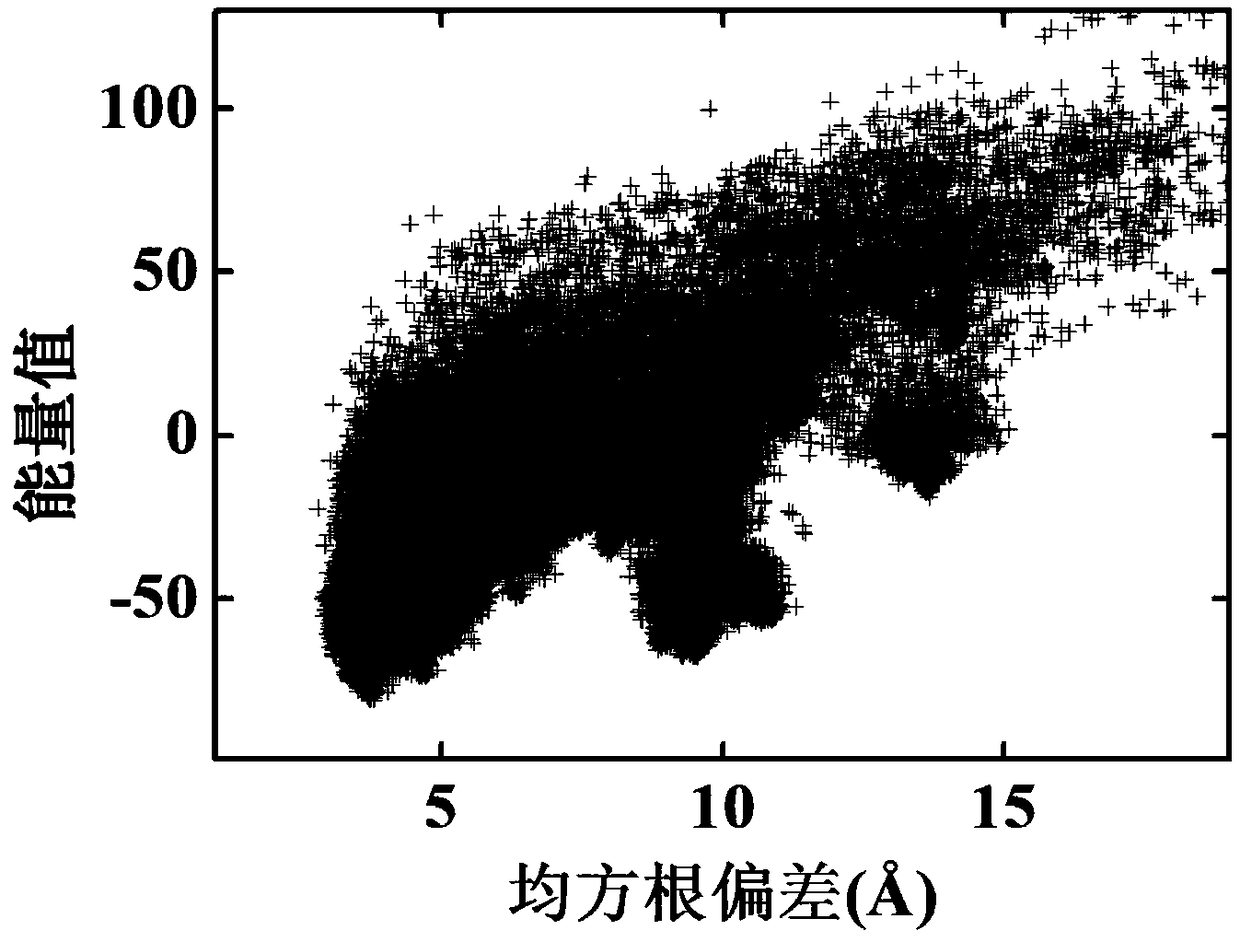
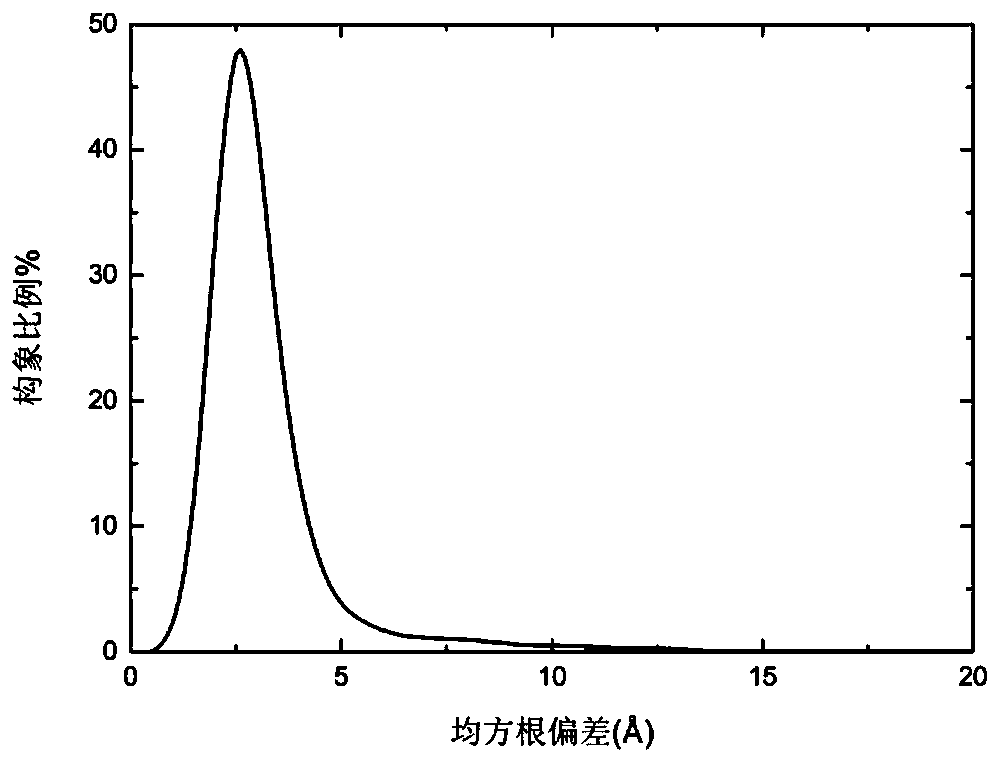
Multi-modal protein structure prediction method based on crowding-out strategy
ActiveCN109360601AAlleviate imprecisionIncreased complexityBiostatisticsInstrumentsLocal optimumDiagnostic Radiology Modality
A multi-modal protein structure prediction method based on a crowding-out strategy comprises the following steps of 1) giving input sequence information and a protein force field model; 2) initializing; 3) carrying out mutation and crossover operation; 4) generating an archiving set; 5) calculating a cluster center and a cluster radius; 6) carrying out crowding-out operation; 7) carrying out clustering operation; and 8) determining whether to satisfy a termination condition, if the condition is satisfied, terminating and outputting all optimal solutions. By using the multi-modal protein structure prediction method based on the crowding-out strategy, under the framework of a differential evolution algorithm, the crowding-out strategy is used to adaptively form multiple modals in an evolution process, all the local optimum solutions of the model can be discovered, and the local optimum solutions can be stored as many as possible in the process so that the prediction precision of the protein structure prediction method can be increased. By using the multi-modal protein structure prediction method based on the crowding-out strategy, the prediction precision is high.
Owner:ZHEJIANG UNIV OF TECH
Multi-variation strategy protein structure prediction method combined with evaluation of exclusion degree
ActiveCN109872770AIncrease diversityAlleviate the problem of low sampling efficiencyInstrumentsMolecular structuresAlgorithmMetapopulation
The invention relates to a multi-variation strategy protein structure prediction method combined with evaluation of exclusion degree. The multi-variation strategy protein structure prediction method includes the steps: under an evolution algorithm framework, 1) establishing three different variation strategies, selecting one variation strategy according to a mode of roulette to perform variation on conformation, and performing 3-segment assembling for once time on the generated variation conformation to generate a variation conformation; 2) performing crossed operation on the variation conformation; and 3) using a Rosetta energy function score3 and a Monte Carlo Boltzmann acceptance criteria to select the conformation with the index {1, 2, ..., NP / 2}, and using an exclusion index Exclusion, and the Monte Carlo Boltzmann acceptance criteria for selecting the conformation with the index {(NP / 2)+1, (NP / 2)+2,..., NP}. The multi-variation strategy protein structure prediction method combined with evaluation of exclusion degree can increase the diversity of populations, and can alleviate the problem that the energy function is not accurate, thus improving the sampling efficiency. The multi-variation strategy protein structure prediction method combined with evaluation of exclusion degree has high sampling efficiency and high prediction accuracy.
Owner:ZHEJIANG UNIV OF TECH
A population protein structure prediction method based on fragment resampling
ActiveCN109086566AImprove search efficiencyIncrease diversitySpecial data processing applicationsProtein targetEnergy functional
A method for predicting population protein structure based on fragment resampling is disclosed. Firstly, Rosetta is used to generate candidate conformers, and a part of conformers with lower energy isselected from the total candidate conformers as the initial population. Then, the population was cross-manipulated to exchange the Loop region of individual population, and the individuals with low energy were selected as the next generation population by using the energy function of individual population. Secondly, the average residue pair distance fraction combined with clustering method was used to select high quality fragments. Finally, using the newly constructed fragment library, the final three-dimensional structure of the target protein can be obtained by assembling the fragments again. The invention provides a population protein structure prediction method based on fragment resampling with high prediction accuracy.
Owner:深圳新锐基因科技有限公司
Vitamin B12 and BtuF protein interaction analysis method
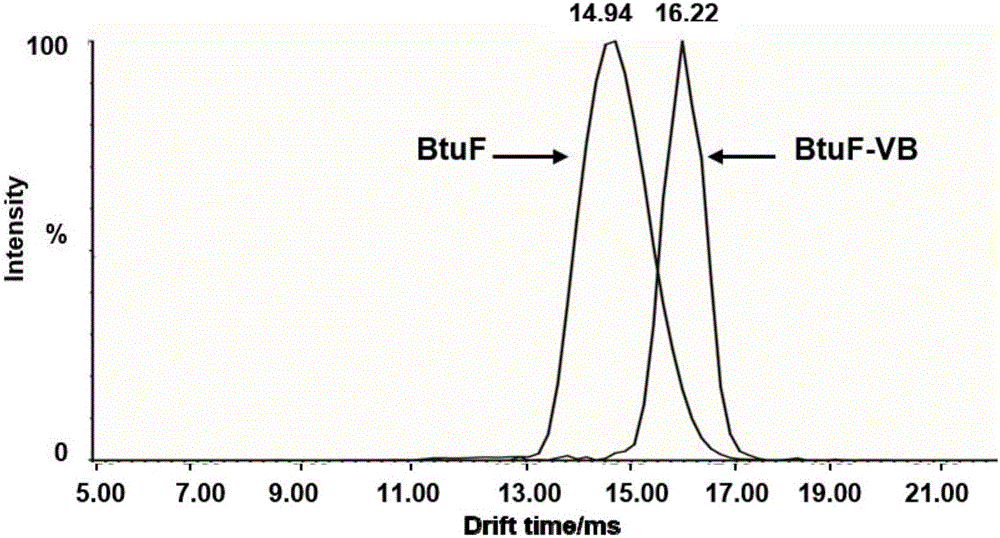
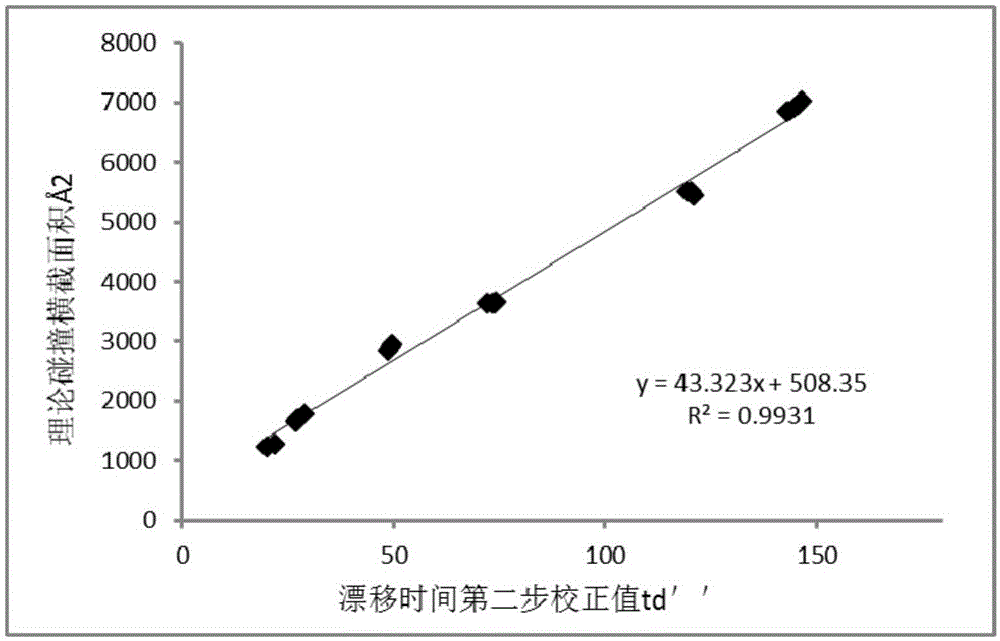
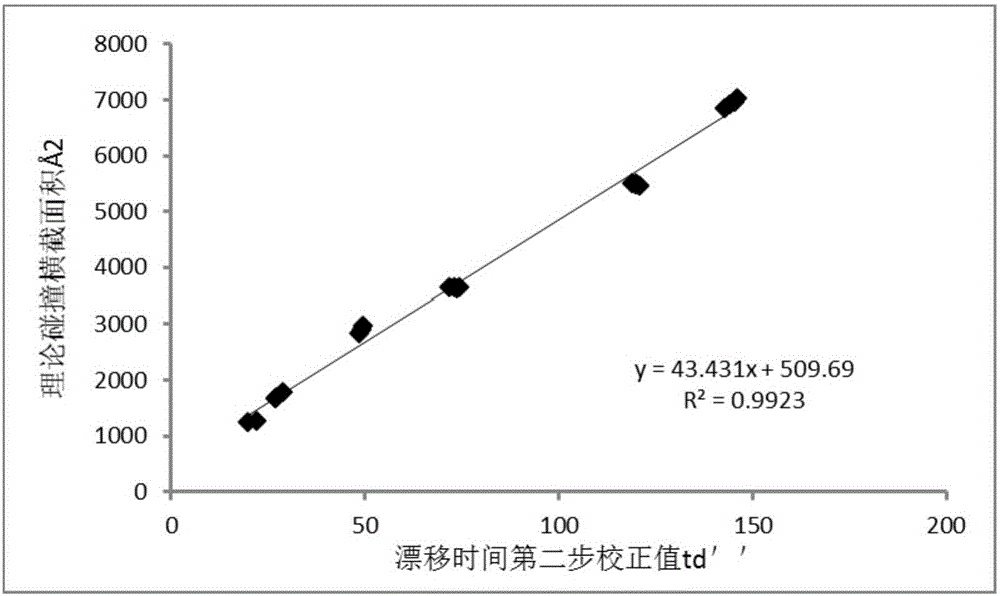
ActiveCN106770613ALow purity requirementEasy to operateMaterial analysis by electric/magnetic meansVitamin B12Mass Spectrometry-Mass Spectrometry
The invention discloses a vitamin B12 and BtuF protein interaction analysis method. According to the method, BtuF protein is replaced into pH8.0 ammonium acetate buffer solution, vitamin B12 is added according to the mole ratio of 1:1 to prepare BtuF-VB solution, the BtuF protein, a BtuF-VB protein compound and a protein standard substance are subjected to mass spectrometric analysis under the same mass spectrum condition, ion mobility data of the BtuF protein, the BtuF-VB protein compound and the protein standard substance are acquired and processed to obtain respective drift time distribution, the collision cross section area of the BtuF and the BtuF-VB is acquired according to drift time distribution and the theoretical collision cross section area of the protein standard substance and drift time distribution of the BtuF protein and the BtuF-VB, BtuF-VB interaction is analyzed according to drift time distribution and the collision cross section area, and the vitamin B12 and the BtuF protein are better in structural uniformity and more compact in conformation after combination. The ion mobility mass spectrometry method is simple in operation, less in needed sample amount and good in repeatability, and can rapidly and accurately provide protein structure information.
Owner:NANJING UNIV OF SCI & TECH
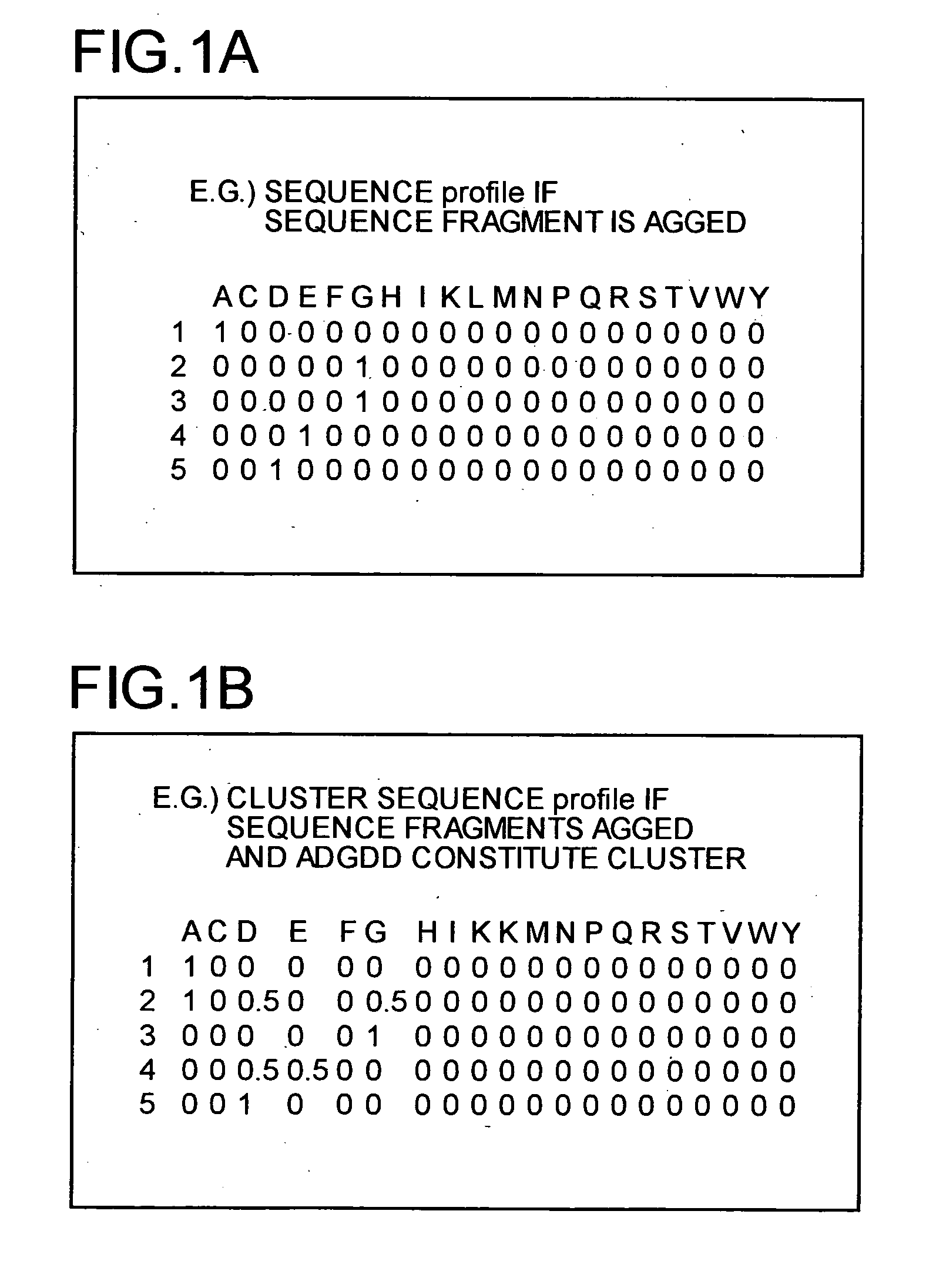
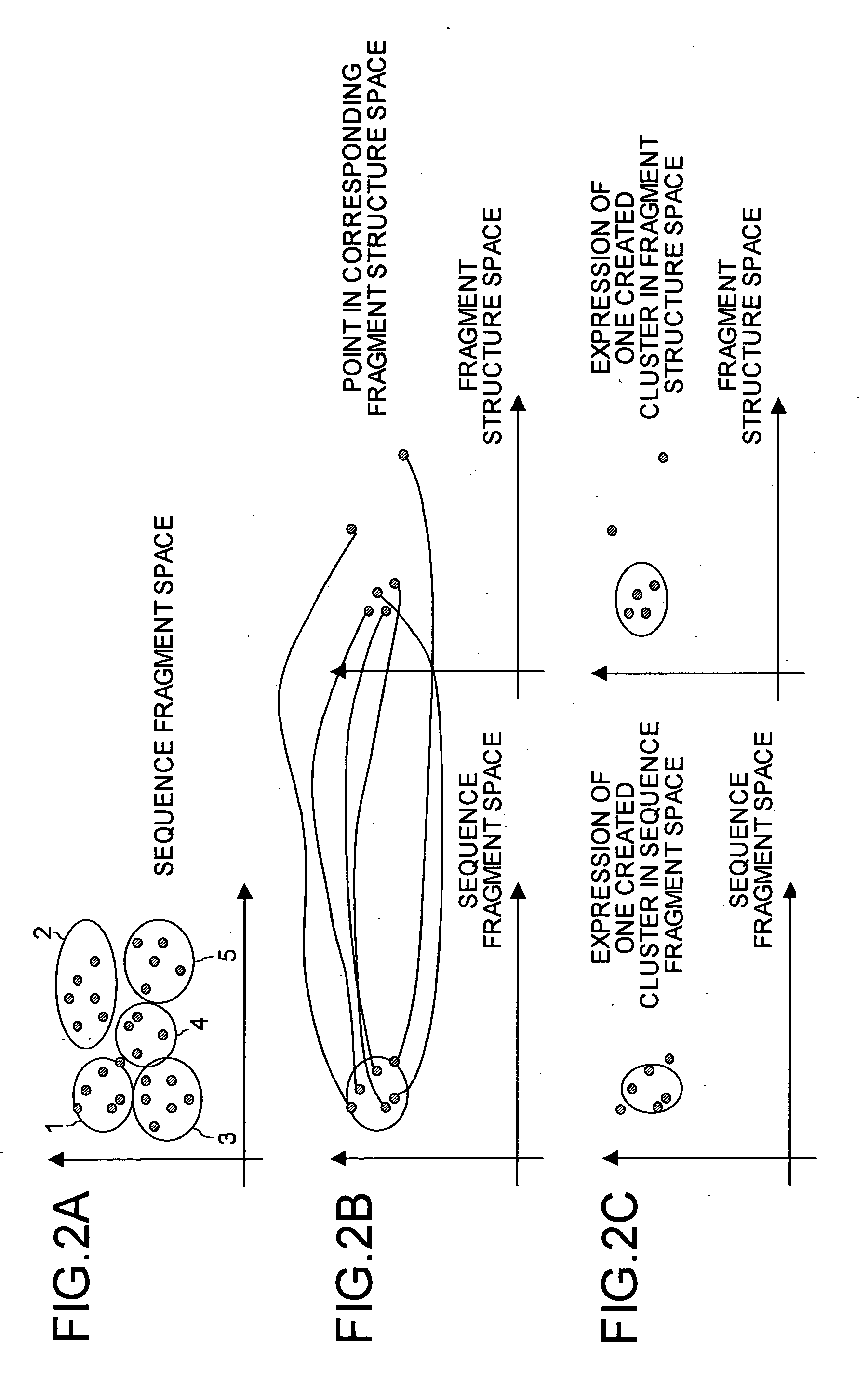
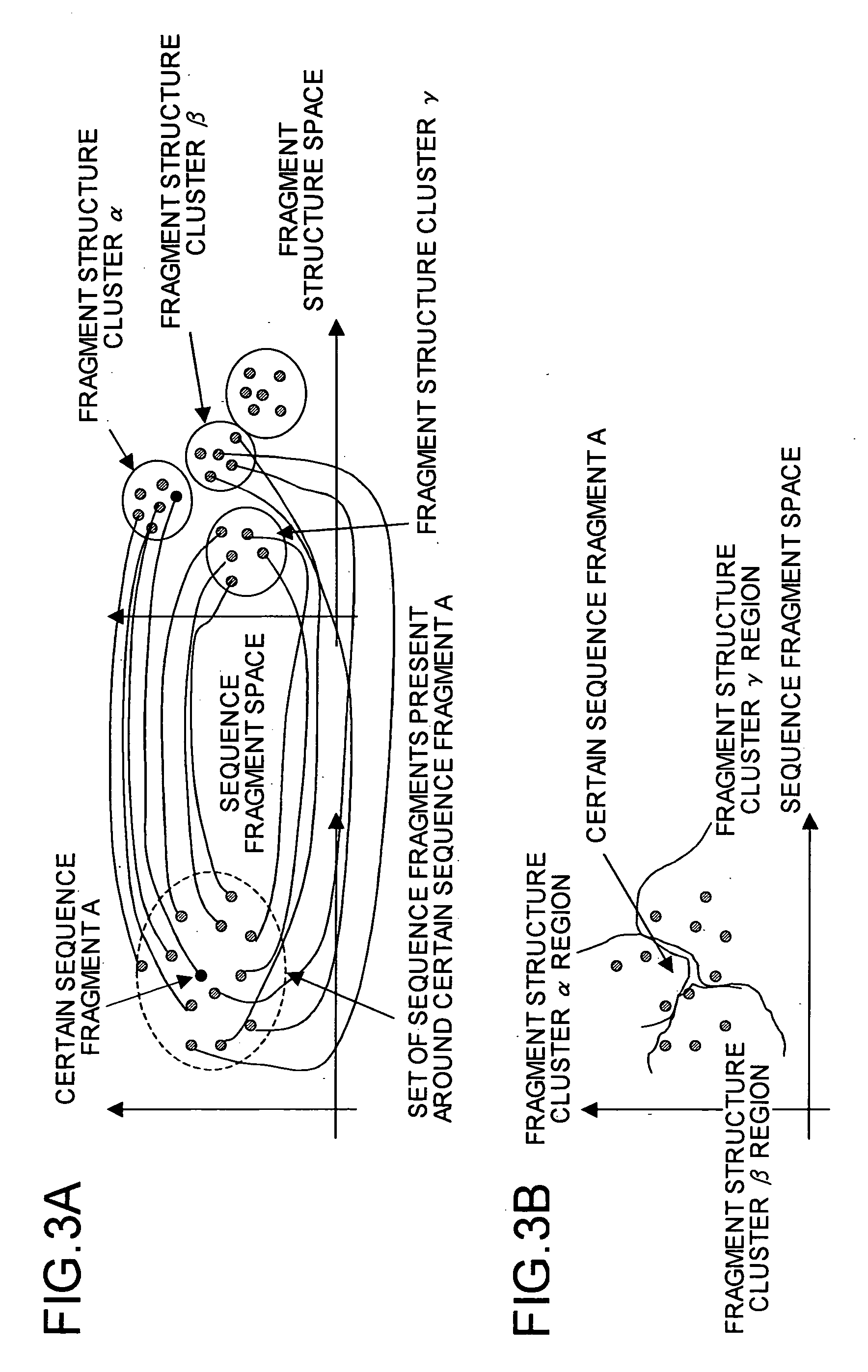
Protein structure prediction device, protein structure prediction method, program, and recording medium
According to a protein structure prediction device, which structure cluster a sequence present around a sequence A and resembling the sequence A belongs to in a structure space (which structure cluster a sequence belongs to when the sequence resembles in what way) is determined by calculation, and a virtual cluster is created around the sequence. When an unknown structure sequence fragment X is given, information on whether the fragment resembles sequence A or C is collected, virtual clusters are combined depending on the information, and a which structure cluster the sequence belongs to is predicted finally.
Owner:CELESTAR LEXICO SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com