Protein structure prediction method based on residue contact information crossover strategy
A protein structure and prediction method technology, applied in the field of protein structure prediction based on residue contact information cross strategy, to achieve the effect of improving prediction accuracy and search efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0034] The present invention will be further described below in conjunction with the accompanying drawings.
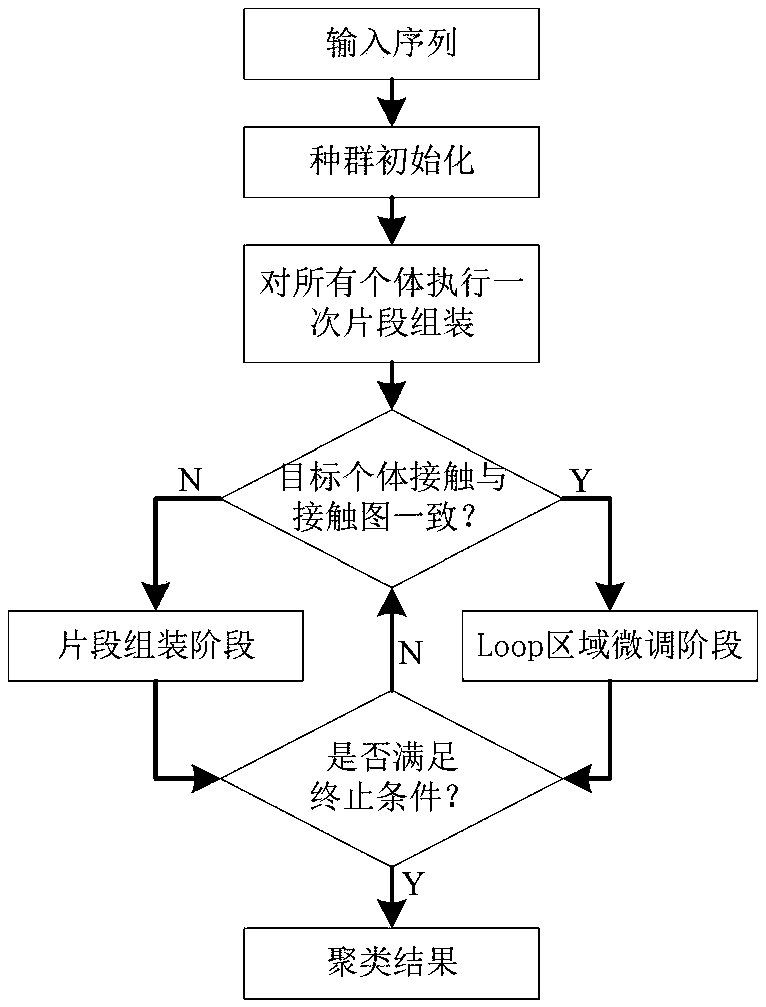
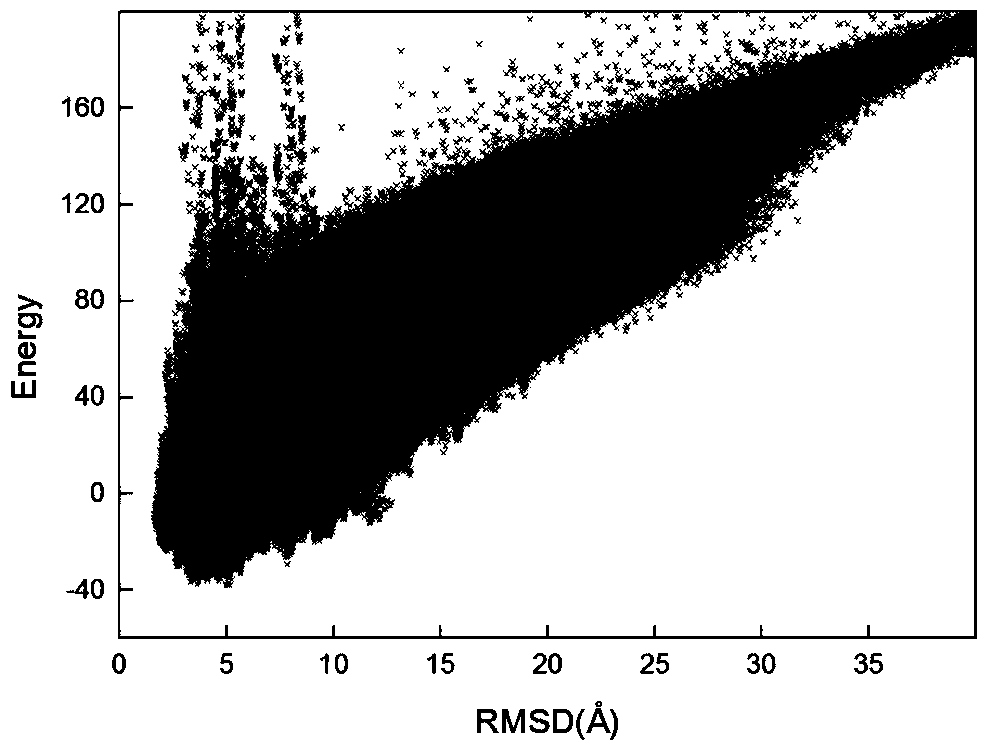
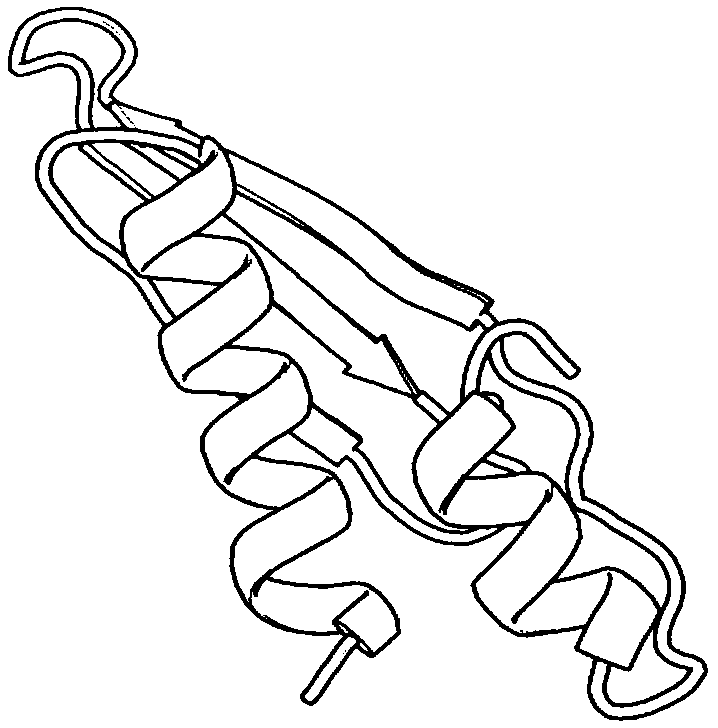
[0035] refer to Figure 1 ~ Figure 3 , a protein structure prediction method based on residue contact information intersection strategy, including the following steps:
[0036] 1) Given the input sequence information, use the Robetta server to obtain the fragment library of the sequence;
[0037] 2) Use RaptorX-Contact to predict the contact information of the sequence, and record the residue pairs whose contact probability is greater than 0.6, assuming that the contact probability of N residue pairs meets this condition, and record the contact between the kth residue pair, Contact means Cα-Cα Euclidean distance The exposure probability is denoted as P k , satisfying P k >0.6, k∈{1,...,N}, and record the residue index number i of the kth residue pair k and j k , and satisfy i k k ;
[0038] 3) Initialization: population size NP, according to the input sequence, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com