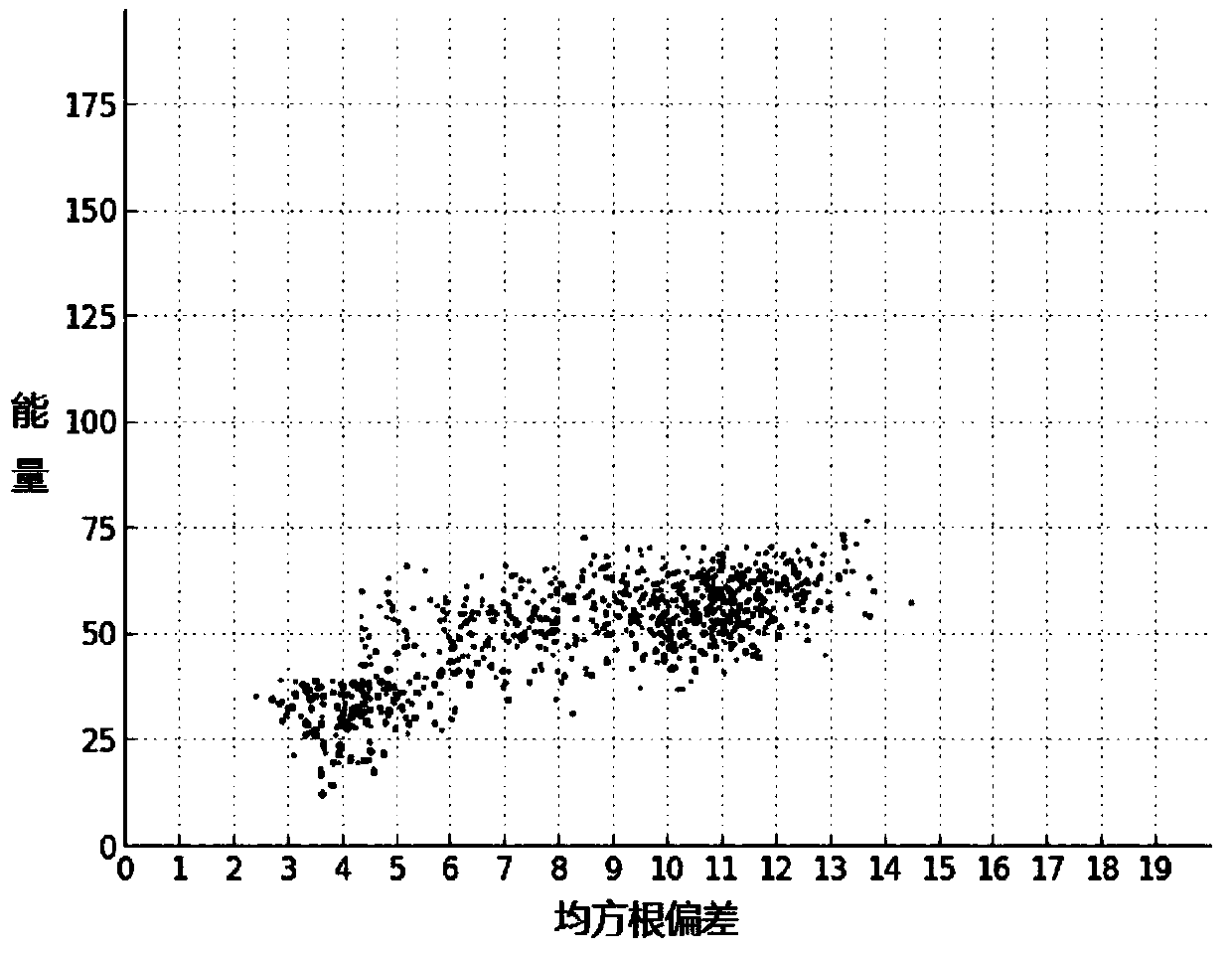
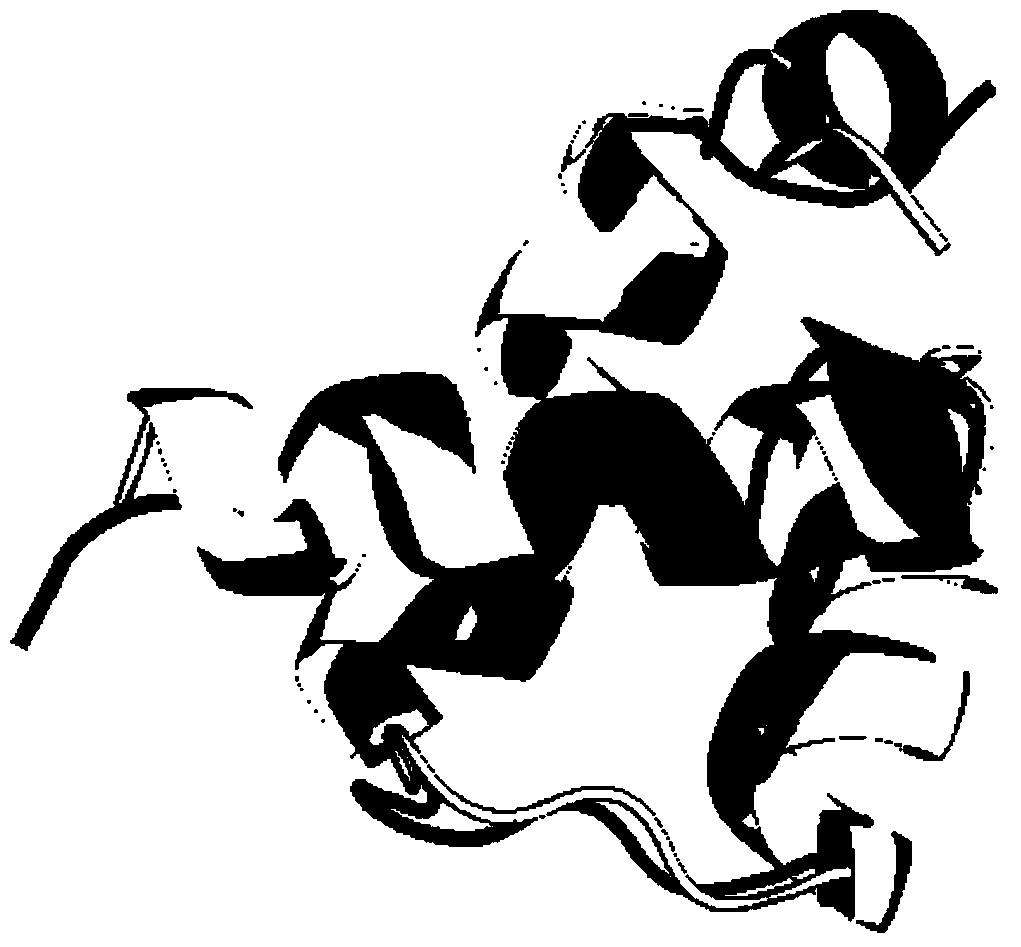Protein structure prediction method based on tree structure replica exchange and fragment assembly
A protein structure and prediction method technology, applied in the field of three-dimensional structure prediction of protein structure, can solve problems such as inaccurate energy model, difficult protein folding problem, and huge protein conformation space
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0059] The present invention will be further described below in conjunction with the accompanying drawings.
[0060] refer to figure 1 and figure 2 , a method for protein structure prediction based on tree-structured copy exchange and fragment assembly
[0061] , the prediction method includes the following steps:
[0062] A1. For protein conformation processing, the ID number is 1ENH, and its sequence is RPRTAFSSEQLARLKREFNENRYLTERRRQQLSSELGLNEAQIKIWFQNKRAKI. The process is as follows;
[0063] STEP1.1. According to the obtained protein amino acid sequence sequence, use the Rosetta package software pose_from_sequence function to construct a long protein chain;
[0064] STEP1.2, and use the Mover object SwitchResidueTypeSetMover built by Rosetta to obtain the long protein chain, and use its apply method to convert the all-atom conformation of the long protein chain into the bone chain atomic conformation. The protein conformation is represented by pose, which is always red...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com