Protein structure predicating method based on tree search and fragment assembly
A protein structure and prediction method technology, applied in the field of computer applications, can solve the problems of high time cost, huge calculation amount, and many computer resources, and achieve the effect of improving the convergence ability and reducing the complexity of the search space.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0037] The present invention will be described in detail below in conjunction with the accompanying drawings.
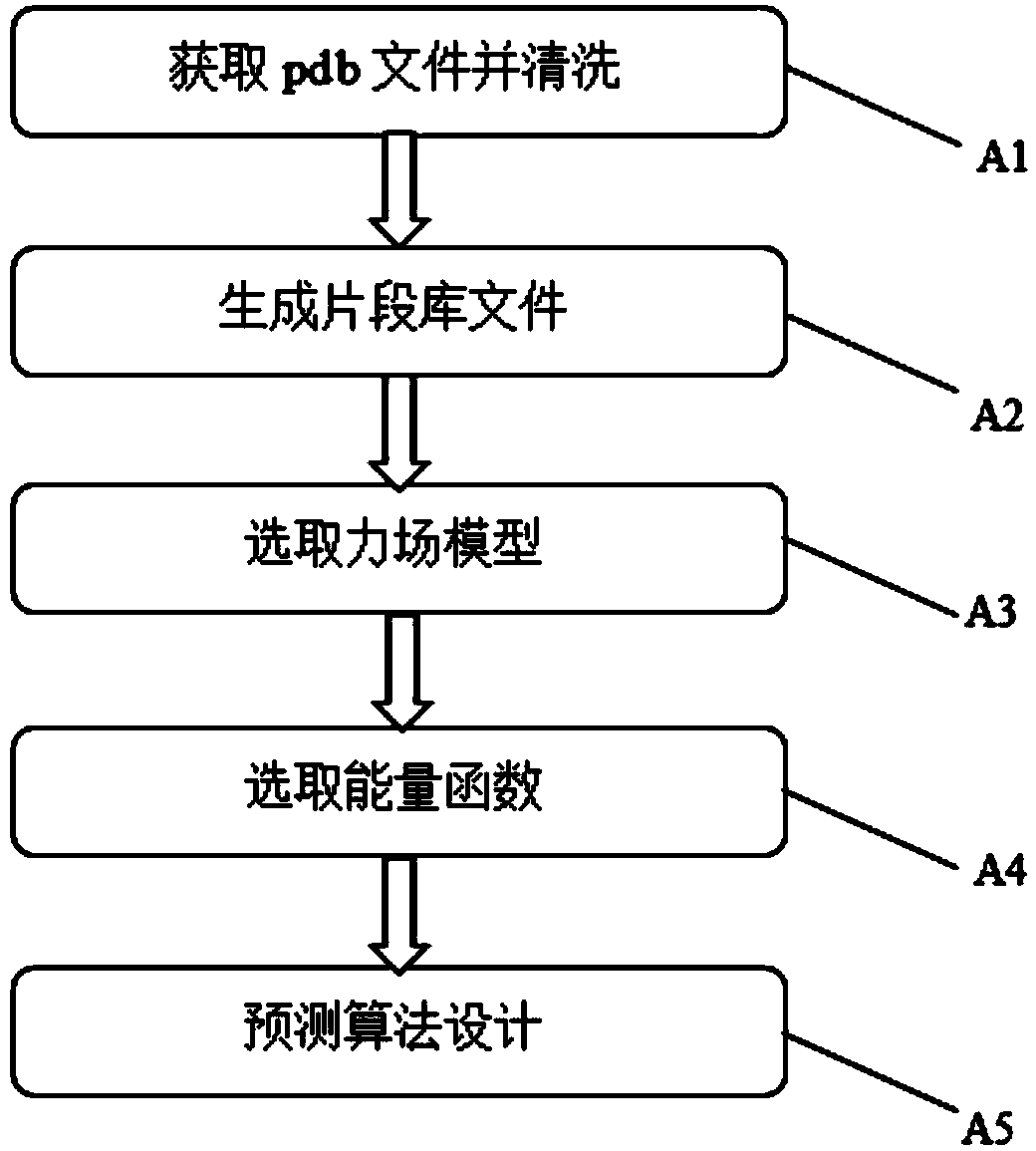
[0038] refer to figure 1 , a protein structure prediction method based on tree search and fragment assembly, which consists of the following steps:
[0039] A1. Obtain the pdb format file of the protein and clean the required data. Download the required protein pdb file from the RCSB official website. Since the pdb format file contains a lot of information that is not needed in the prediction, it needs to be further "cleaned" before it can be used for the next step of structure prediction. The method of "cleaning" the pdb file: use the python scripting language to write a script program, select the information in the pdb file that contains the detailed information of protein atoms, intercept it and save it as a new pdb file.
[0040] A2. Download the generated fragment library from the relevant website.
[0041] A3. Select the force field model. The Rosetta forc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com