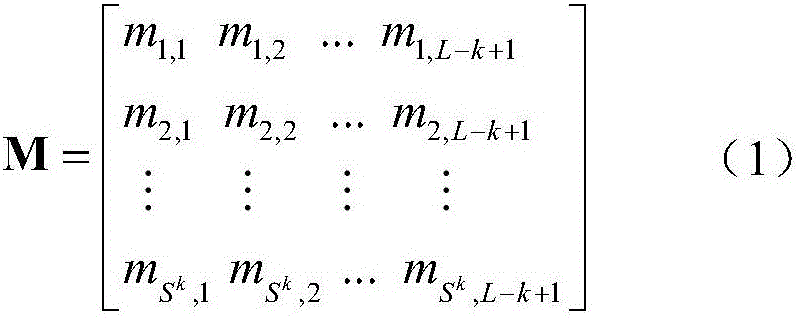Sequence-order dependent frequency matrix-based biological sequence evolution information extraction method and application thereof
A biological sequence, frequency matrix technology, applied in the field of bioinformatics, can solve the problem of inability to extract adjacent position dependent information
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
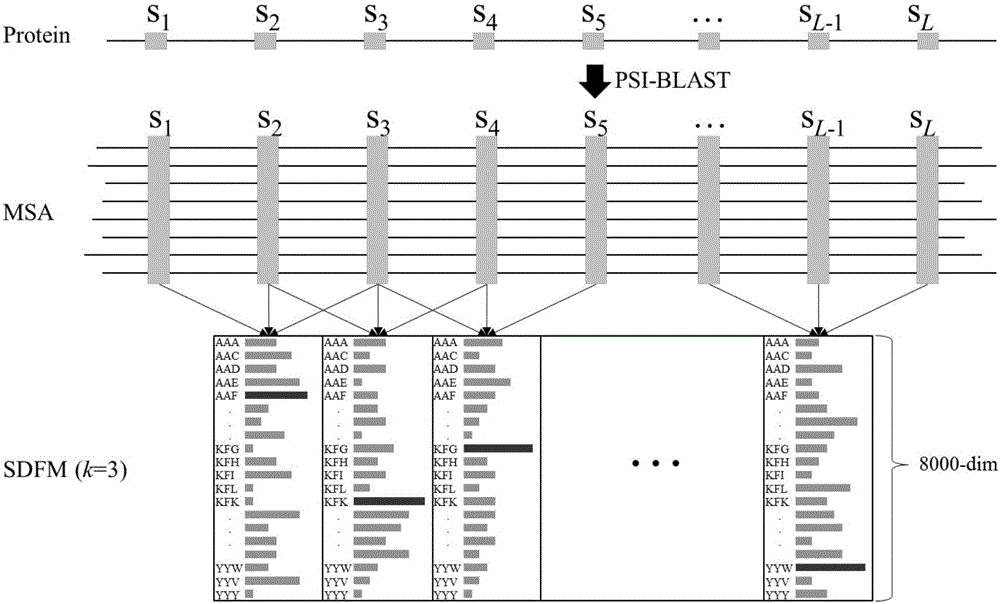
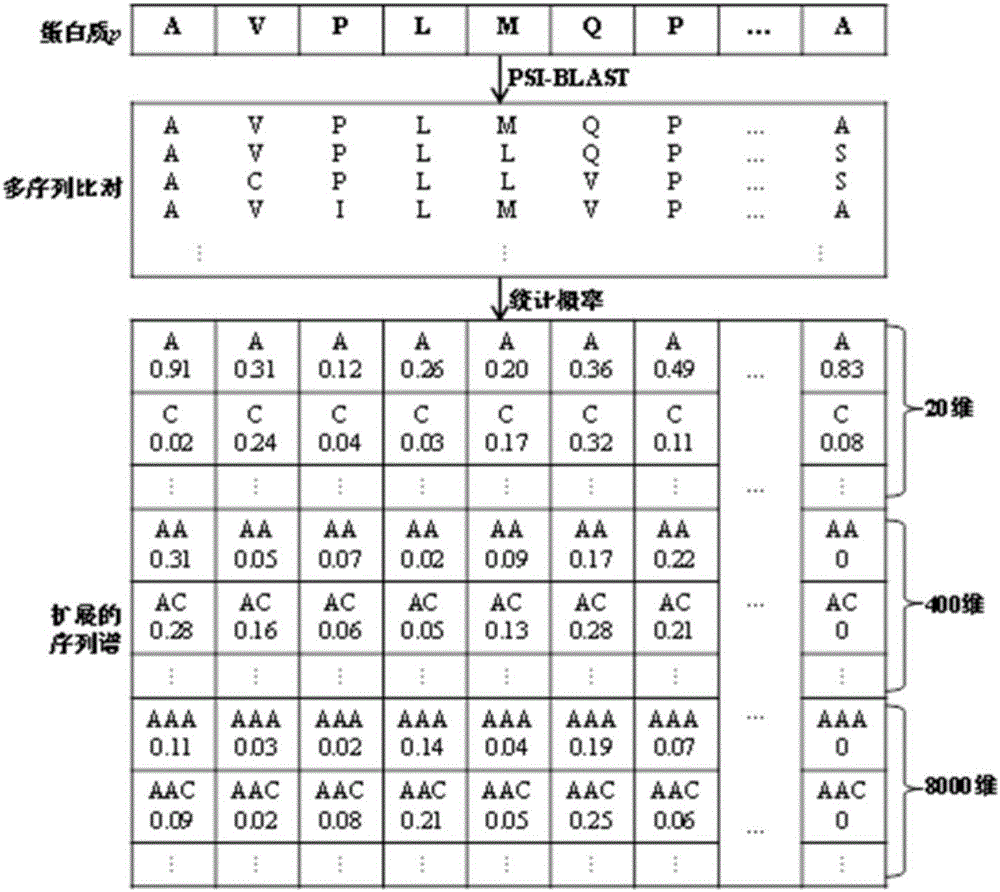
[0034] Taking a protein sequence as an example, for any protein sequence, first use a sequence alignment tool such as PSI-BLAST to search a large-scale protein database such as NRdb90 to obtain the multiple sequence alignment MSA of the query protein. Then the frequency of amino acid sequence substrings with a length of 3 appearing at each position in the multiple sequence alignment MSA was counted. Such as figure 1 As shown in , the histogram in each column represents the probability distribution of the occurrence of amino acid substrings at this position, and the label of each row is the type of amino acid substrings. For a protein of length L, generate a corresponding SDFM of size 20 k ×(L-2). The process of generating sequence-dependent frequency matrix k=3 of protein sequence is as follows: figure 1 shown.
[0035] When only counting the occurrence probability of biological substrings with a length of 1 in multiple sequence alignments, that is, k=1 in formula (1), the...
Embodiment 2
[0037] On the basis of Example 1, we can combine multiple SDFMs generated based on biological sequence substrings of different lengths to include more biological sequence evolution information. Taking protein SDFM as an example, we can combine SDFM with k=1, 2, 3 into a matrix. The schematic diagram of the combination of the sequence-dependent frequency matrix of a specific protein sequence is as follows figure 2 shown. Firstly, SDFMs of biological substrings of different lengths were generated, and then aligned and spliced according to the corresponding amino acid positions to form a matrix with a larger dimension.
[0038]The technical scheme of the present invention takes into account the interdependence between biological sequence sites, and increases the information of biological sequence site dependencies on the basis of the original specificity scoring matrix PSSM, and uses the sequence-dependent frequency matrix SDFM to perform biological Sequence evolution informa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com