Selection of embryo of test tube baby through sequencing by single cell genome of polar body or embryo
A genome and single-cell technology, applied in the direction of microbial measurement/testing, biochemical equipment and methods, etc., can solve problems such as inability to diagnose
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0121] The following examples facilitate a better understanding of the present invention, but do not limit the present invention. The experimental methods in the following examples are conventional methods unless otherwise specified. The test materials used in the following examples, unless otherwise specified, were purchased from conventional biochemical reagent manufacturers. Quantitative experiments in the following examples were all set up to repeat the experiments three times, and the results were averaged.
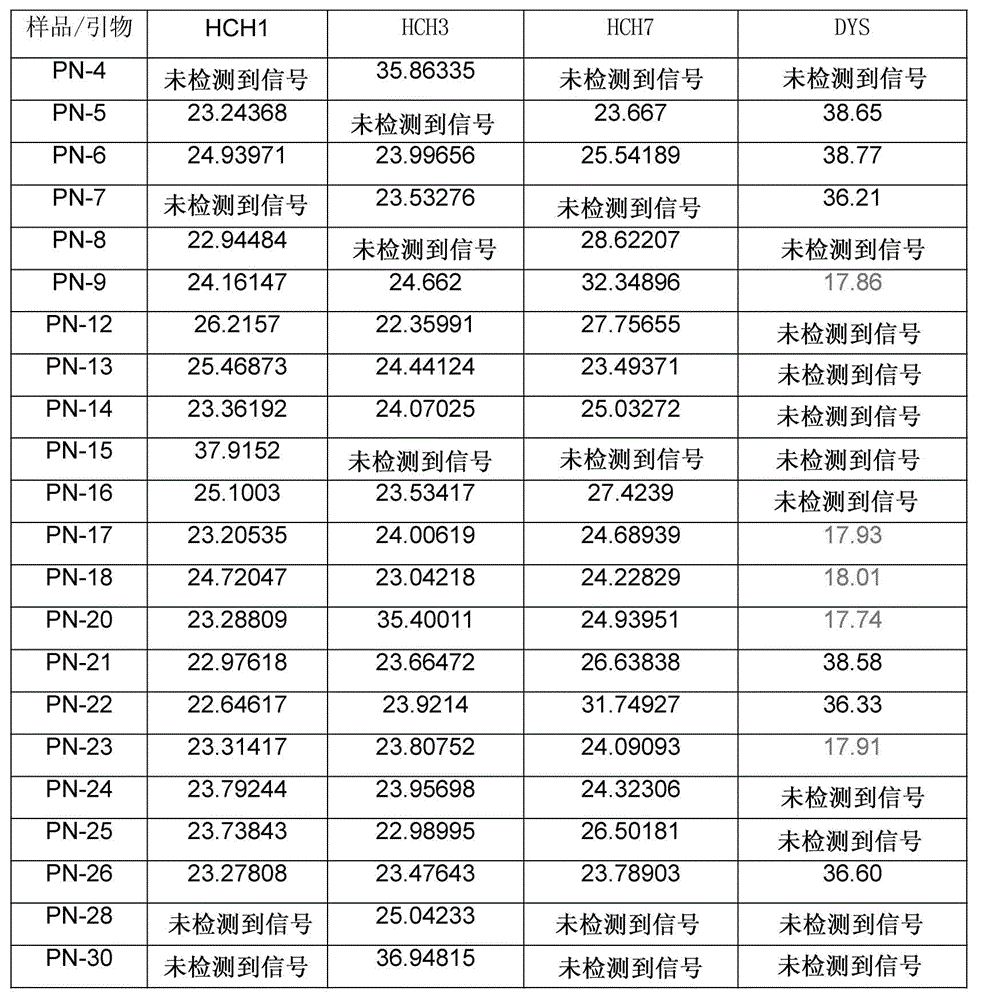
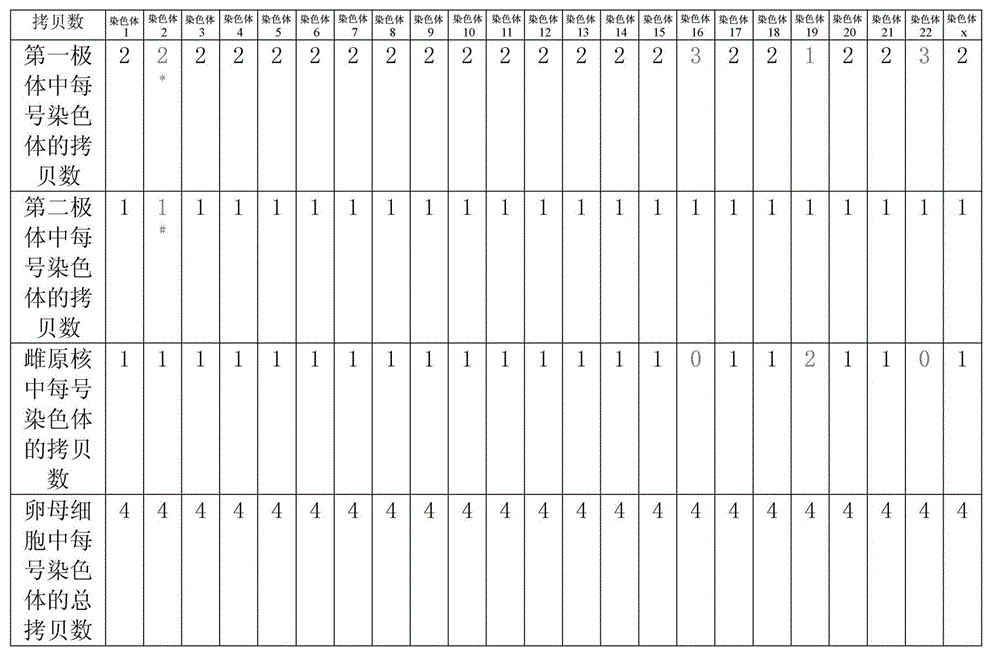
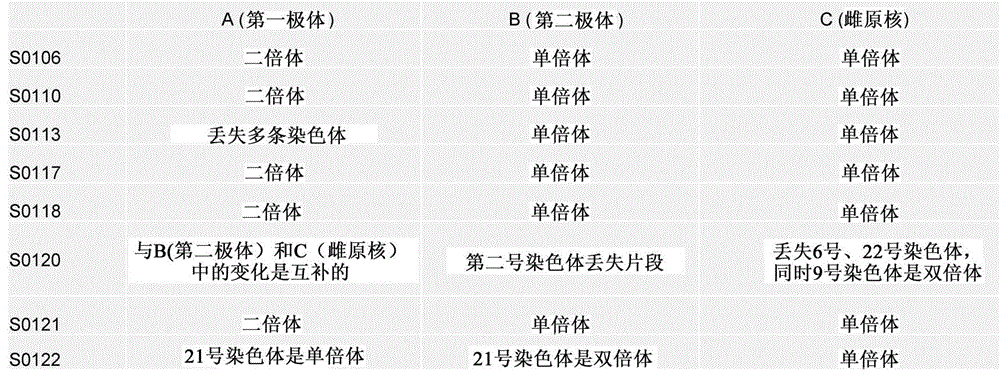
[0122] Part 1: A method for genome sequencing using the first polar body and the second polar body and the female pronucleus of the embryo, including the following steps:
[0123] 1. The process of obtaining the first polar body, the second polar body and the female pronucleus of the oocyte:
[0124] At 1.0: microinjection of single sperm (ICSI) into eggs at stage MII, put the eggs into (G-MOPS) operating fluid, transfer to the platform of the micromanipulator, and p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com