Method for detecting tumor single cell somatic mutation
A somatic cell mutation and single-cell technology, applied in biochemical equipment and methods, microbial measurement/testing, etc., can solve the problems affecting the detection of tumor single-cell somatic cell mutations, random and systematic errors, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
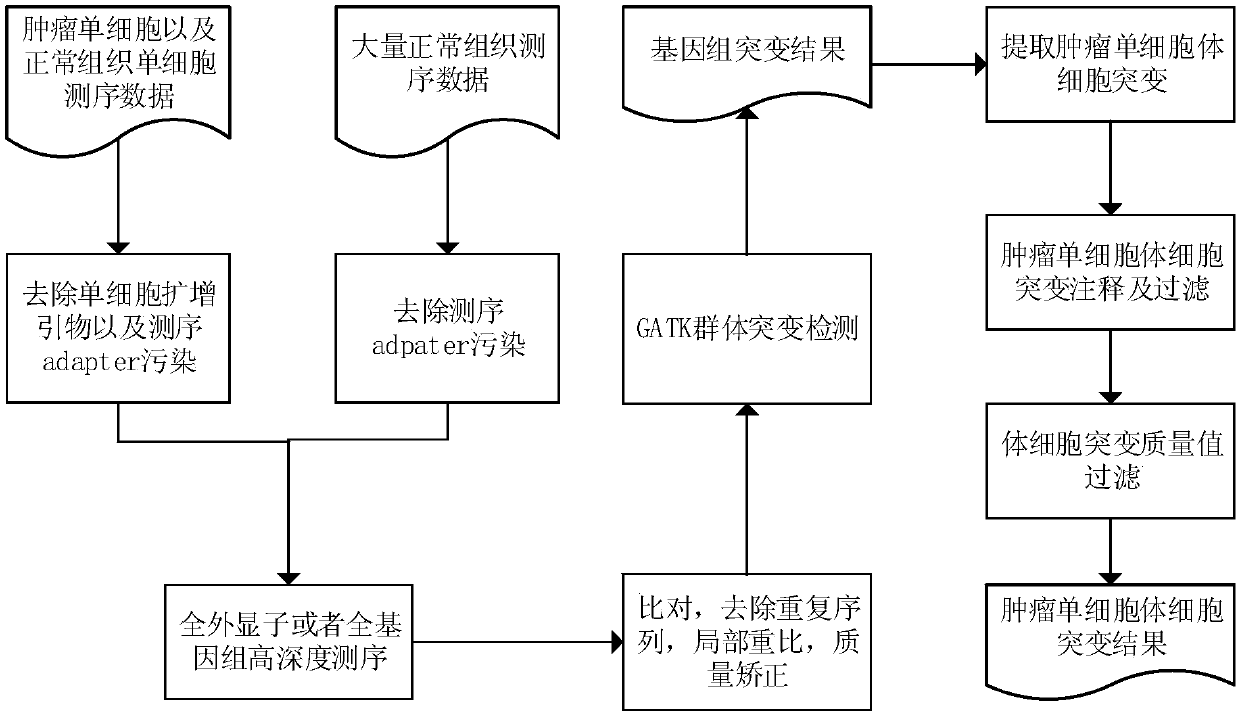
[0032] The following is a more detailed description of the implementation of the present invention, in which the parameters and specific implementation details are used to explain the feasibility and implementation effect of the present invention, and do not constitute a limitation of the present invention.
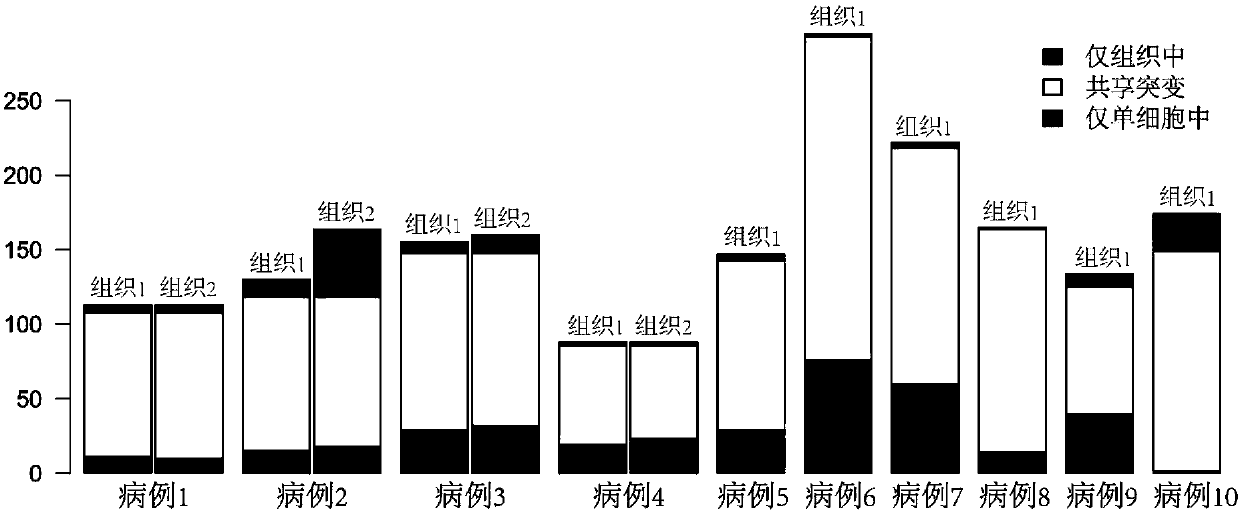
[0033]In this example, the circulating tumor cells, single white blood cells, routinely sequenced blood bulk control and tumor tissue control of ten patients with small cell lung cancer were used as samples, and the somatic mutation of tumor single cells was studied based on next-generation sequencing technology .
[0034] 1. Sample requirements and single cell amplification and sequencing
[0035] The samples were circulating tumor cells from ten cases of small cell lung cancer (the number of circulating tumor cells ranged from 4 to 25 for each case), corresponding monoleukocytes (one for each case), and corresponding blood cells (one for each case) One sample per case)...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com