Trace and complex DNA amplification method
A complex and trace technology, used in complex DNA amplification and trace fields, can solve the problems of amplification failure, non-specific amplification, loss, etc., and achieve the effect of good specificity, good consistency, and reduced tripping.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
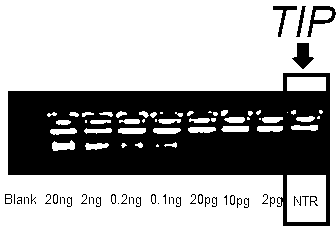
[0045] Example 1, Amplification of trace and complex DNA above 1ng
[0046] Step 1, whole genome DNA denaturation
[0047] Preparation buffer A: including 400 mmol / L KOH, 10 mmol / L EDTA
[0048] Preparation buffer N: including 200 mmol / L HCl, 300 mmol / L Tris-HCl (pH 7.5)
[0049] Add 1 μl of freshly prepared buffer A to a pre-cooled 200 μl thin-walled PCR tube, and then add 1 ul of whole genomic DNA (usually 1~5ng, as low as a single human cell or bacterial genomic DNA). After 3 minutes of denaturation (add 1 minute more for each additional 5ng of genomic DNA; if the amount of original DNA is small, reduce the denaturation time or dilute buffer A), add 1 μl buffer N and mix well.
[0050] Step 2, adding trehalose (TRE)
[0051] Quickly add 5 μl of 1.2M TRE, mix and place on ice.
[0052] The preparation method of 1.2M trehalose. Dilute with sterile DNA-free distilled water and then sterilize with a 0.22 micron filter (Millipore). Aliquots can be stored at -20°C for lon...
Embodiment 2
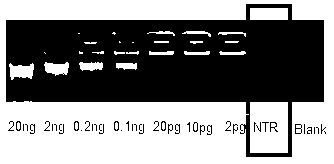
[0066] Example 2, High sensitivity, high fidelity, high uniformity, and high efficiency amplification of trace and complex DNA less than or equal to 0.5ng
[0067] Step 1, denaturation of trace and complex DNA
[0068] Referring to Example 1, for ≤ 0.5ng of pre-purified and extracted trace and complex DNA samples, dilute buffer A and buffer N described in Example 1 with 8 times of water, and refer to the denaturation method described in Example 1, DNA Denature in diluted buffer for 8 min.
[0069] When the sample is less than or equal to 0.1ng, the reduction in denaturation time is about one minute when the added DNA is reduced by one level.
[0070] Step 2, adding trehalose (TRE)
[0071] Quickly add 5 μl of 1.2M TRE, mix and place on ice.
[0072] Step 3, perform amplification
[0073] Then add the WPA reaction mixture described in Example 1 to the reaction system in step 2, turn it upside down several times to mix, add 1 μl of the modified phi29 DNA polymerase (concen...
Embodiment 3
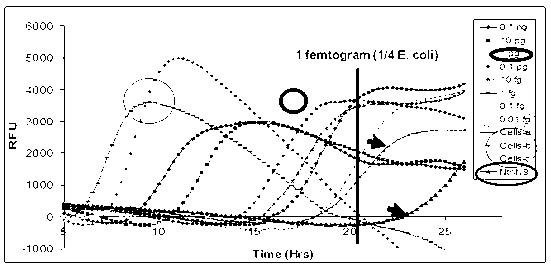
[0076] Example 3, Amplification of Whole Cell DNA from Less Than 10 Cells
[0077] Step 1, Genomic DNA Denaturation
[0078] Preparation of denatured cell lysate: including 400mmol / L KOH, 10mmol / L EDTA, 100mmol / L DTT
[0079] Adjust the volume of cells (including corresponding solutions) to 1 μl, add 1 μl of cell lysate, place on ice, mix, and keep for 5 minutes. Then 1 μl of buffer N was added.
[0080] Step 2, adding trehalose (TRE)
[0081] Quickly add 5 μl of 1.2M TRE, mix and place on ice.
[0082] Step 3, perform amplification
[0083] Then add the WPA reaction mixture described in Example 1 to the reaction system in step 2, turn it upside down several times to mix, add 1 μl of the modified phi29 DNA polymerase (concentration 1 μg / μl), mix and turn to the constant temperature platform, in React at 30°C for 16 hours.
[0084] Step 4, terminate the reaction
[0085] After the amplification is completed, place the PCR reaction tube in a 70°C water bath for 20 minut...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com