Patents
Literature
302 results about "Function genes" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Genes are segments of DNA encoding information that ultimately: direct the production of RNA molecules that serve a variety of functions that include dictating the synthesis of proteins that perform a wide variety of functions in the body. regulating (turning on or turning off) the expression of other genes.
Rice whole genome SNP chip and application thereof
ActiveCN102747138AImprove throughputGood repeatabilityNucleotide librariesMicrobiological testing/measurementManufacturing technologyGermplasm
The present invention discloses a rice whole genome SNP chip and an application thereof. A method for preparing the chip comprises: (1) obtaining a first class of probes on a chip, wherein sequencing is performed to obtain a parental genome sequence, resequencing data of other rice varieties in a public database are combined, a Nipponbare genome is adopted as a reference sequence, a MAQ software is adopted to match and analyze all the sequencing data, and finally a SNP marker is screened; (2) obtaining a second class of probes on the chip, wherein a rice function gene is obtained from the public database, sequence difference reflecting gene function is searched, and a SNP / INDEL probe is designed according to the sequence difference; (3) adopting an infinium chip manufacturing technology to produce a SNP chip; and (4) testing accuracy and application efficiency of the chip. The chip of the present invention can be applicable for rice germplasm resource molecule marker fingerprint analysis, seed authenticity detection, filial generation genotyping, and other related researches.
Owner:先正达集团股份有限公司
Methods and compositions for selective RNAi mediated inhibition of gene expression in mammal cells
InactiveUS20050026286A1Reducing sequenceMicroorganismsMicrobiological testing/measurementMammalMammalian cell
RNAi arrays and methods for using the same are provided. The subject arrays are characterized by having two or more distinct RNAi agents. The arrays find use in methods where cells are contacted with the arrays and the activity of the RNAi agents is determined by evaluating the contacted cells. The subject arrays and methods find use in a variety of applications, such as high throughput loss of function genomic applications.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
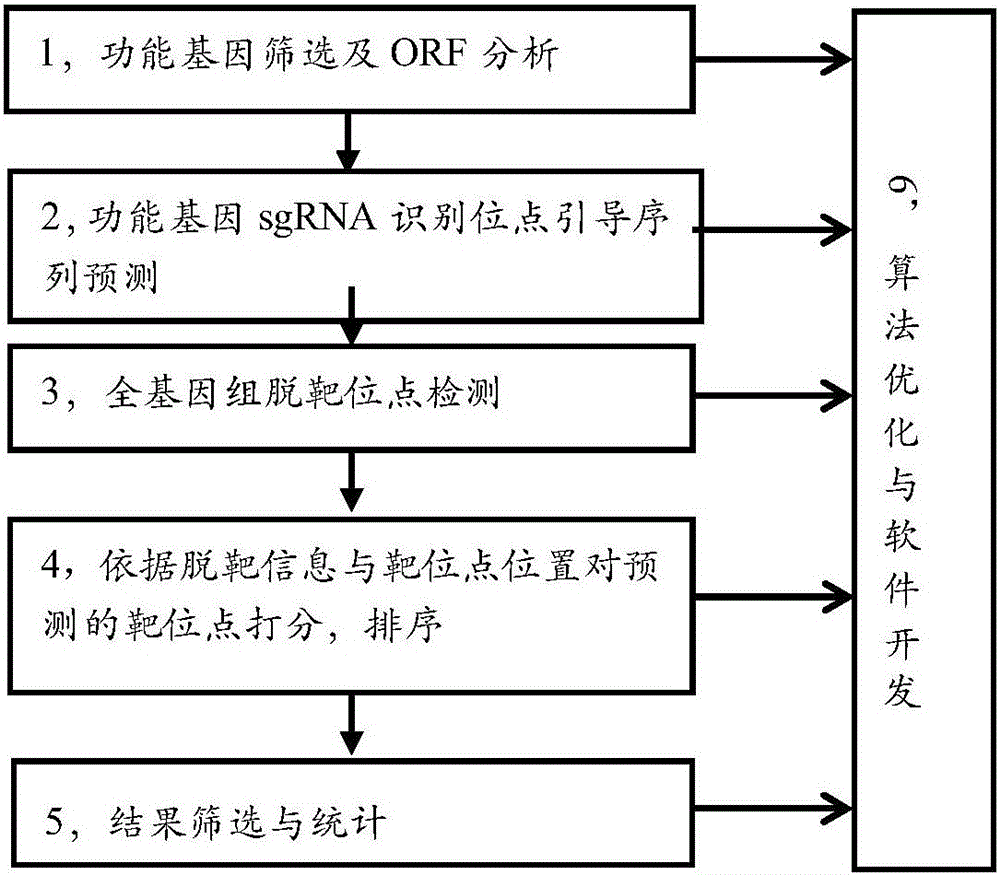
Efficient specific sgRNA recognition site guide sequence for pig gene editing and screening method thereof
ActiveCN105886616AMicrobiological testing/measurementDNA/RNA fragmentationGenome editingScreening method
The invention discloses an efficient specific sgRNA recognition site guide sequence for pig gene editing and a screening method thereof. The screening method includes: screening functional genes, performing ORF analysis, predicting a functional gene sgRNA recognition site guide sequence, detecting whole-genome off-target sites, grading predicted target sites according to off-target information and target site positions, sequencing, screening and statistically counting results, optimizing algorithms and developing software. The efficient specific sgRNA recognition site guide sequence and the screening method thereof have the advantages that the pig specific sgRNA recognition site guide sequence is obtained through strict screening and inspection and includes the sgRNA recognition site guide sequences, for CRISPR-Cas9 gene editing, of all pig protein encoding genes; the authenticating, grading and inspecting algorithms for specific sgRNA recognition and software corresponding to the algorithms and used for predicting and evaluating the pig functional gene sgRNA recognition site guide sequence are widely applicable to the sgRNA specific site prediction of non-model species with whole-genome sequences.
Owner:AGRO BIOLOGICAL GENE RES CENT GUANGDONG ACADEMY OF AGRI SCI
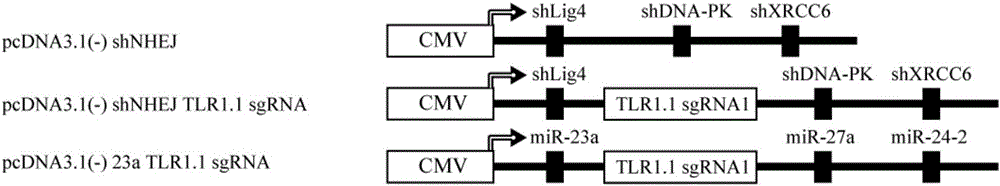
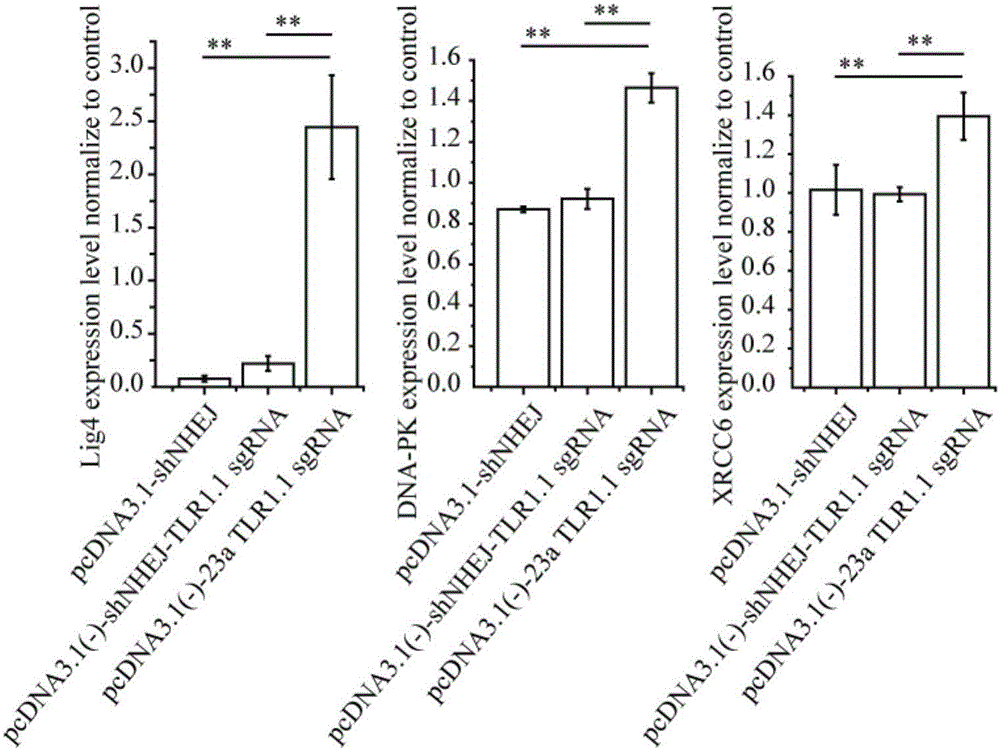
Method for improving efficiency of CRISPR mediated homologous recombination
InactiveCN106399367ANucleic acid vectorVector-based foreign material introductionSignalling pathwaysBiology
The invention discloses a method for improving the efficiency of CRISPR mediated homologous recombination. The method includes the steps of: inhibiting the expression of Lig4, DNA-PK and XRCC6 by shRNA so as to inhibit the non-homologous end joining (NHEJ) repair pathway; conducting fusion expression on sgRNA and shRNA at targeting genome specific positions to form shRNA-sgRNA polycistron; and placing the polycistron at the RNA polymerase II or RNA polymerase III promoter downstream. The carrier constructed by the method provided by the invention has a concise structure and is convenient to use, also enhances the homologous recombination efficiency by dozens of times, greatly expands the function and application of the CRISPR carrier, and is a good tool for functional gene and signal pathway research, medical research, and recombinant protein expression.
Owner:SHENZHEN WEIGUANG BIOLOGICAL PROD
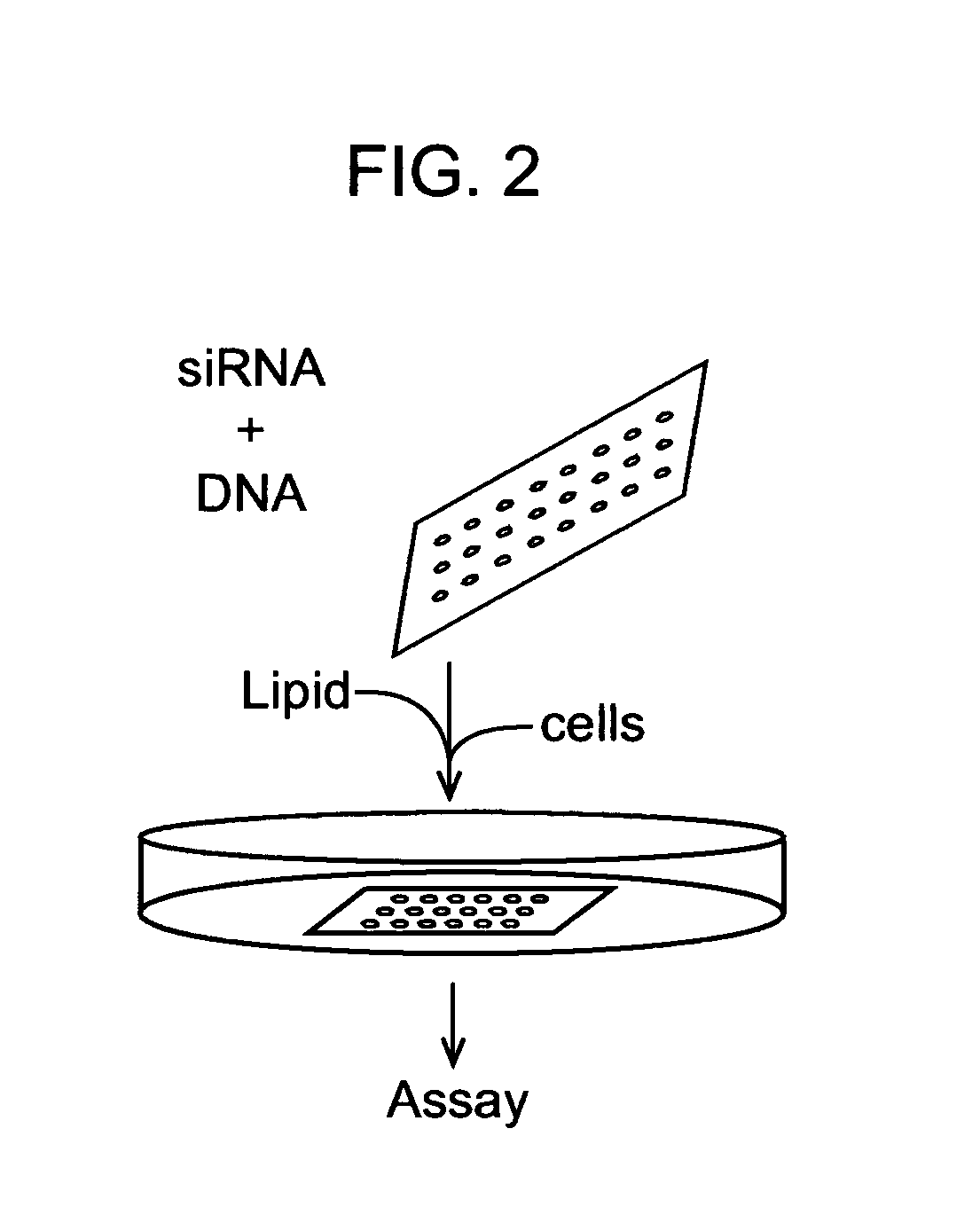
siRNA knockout assay method and constructs
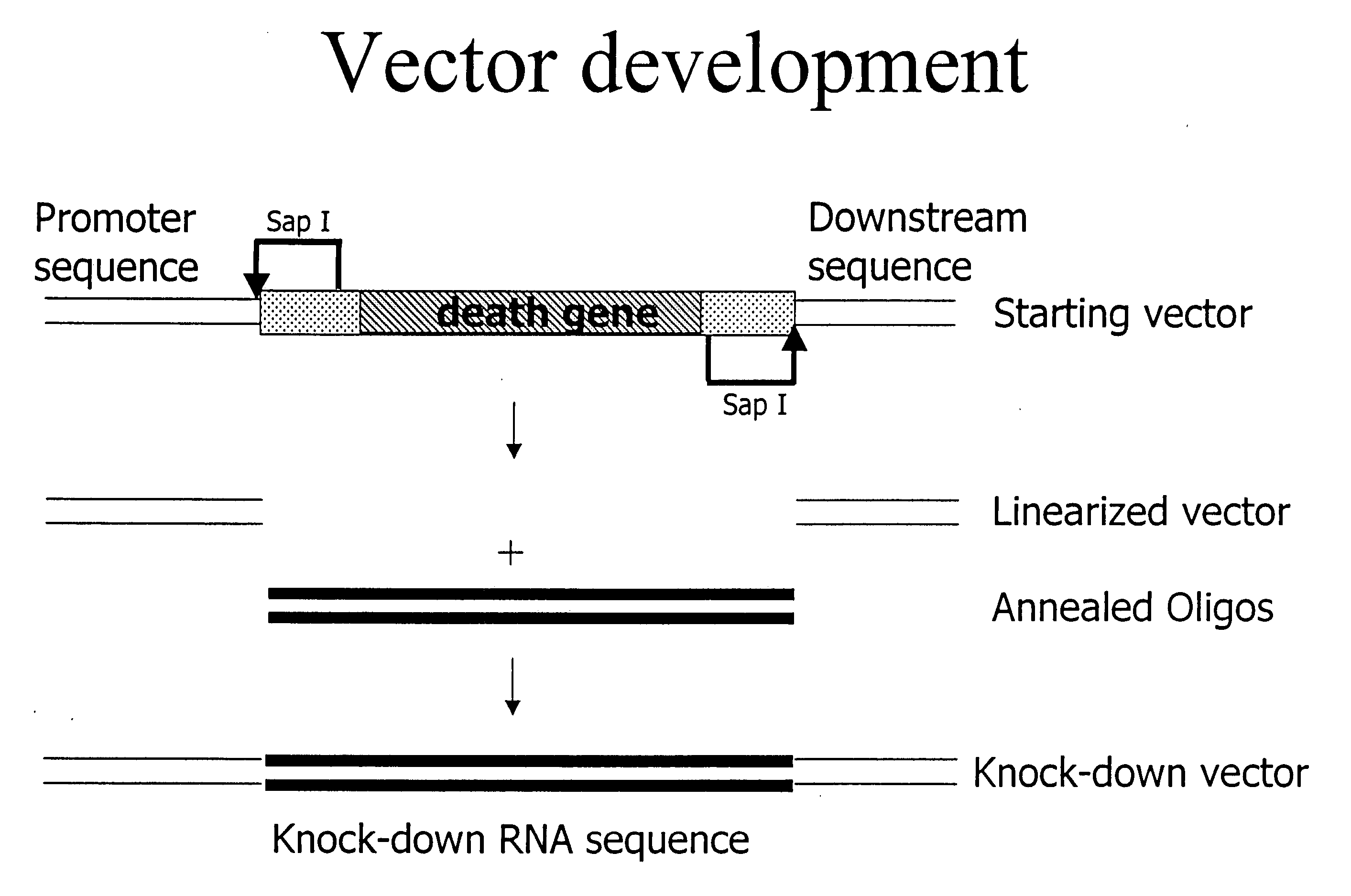
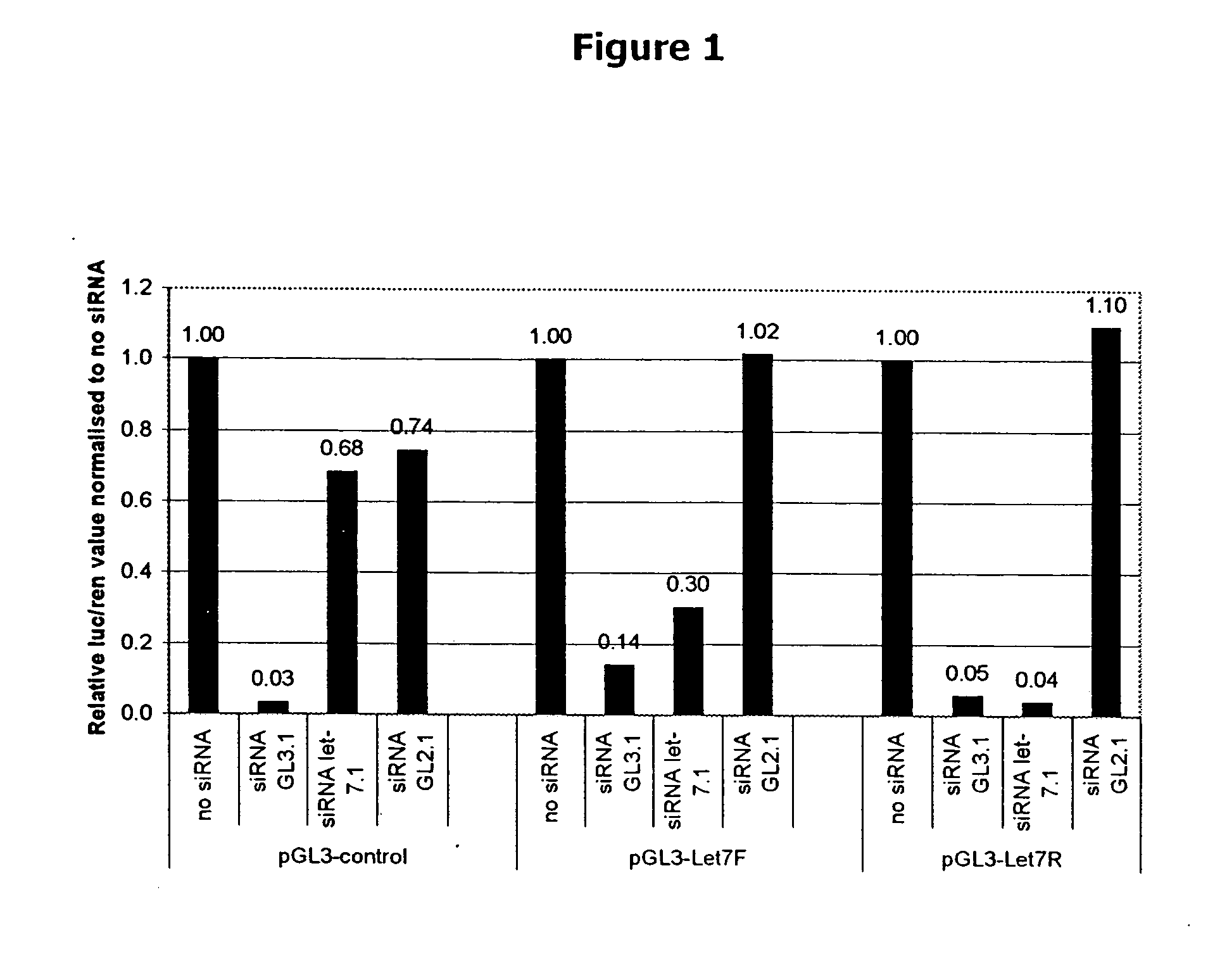
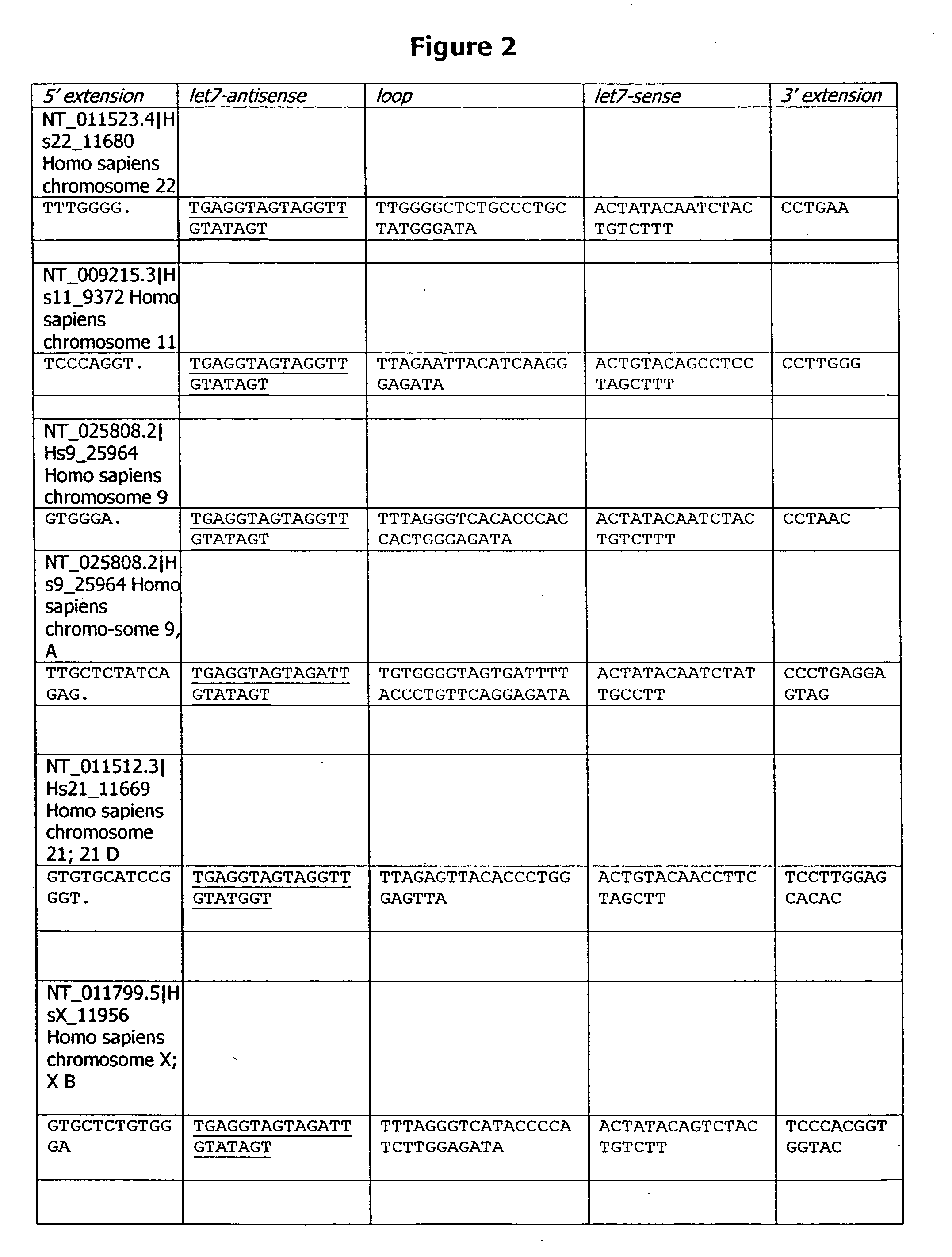
Isolated polynucleotides, and vectors including the same, are disclosed as useful for down-regulation of specific RNA in cells, including a first sequence of about 17 to about 23 nucleotides, complementary to said RNA, and linked to a second sequence capable of forming a loop when said second sequence is RNA. The polynucleotides include self-complementing single-stranded polynucleotides, including a third sequence linked by said second sequence where all nucleotides in said first and said third sequences are complementary. Functional genomic, diagnostic and therapeutic methods are disclosed that involve reducing the amount of a unique RNA sequence in cells using a vector encoding the self-complementing polynucleotide including a first sequence complementary to said RNA sequence. Methods are also disclosed for preparing the polynucleotides, vectors, libraries of vectors, and the temporary knock-down of proteins, such as lethal proteins, during virus or recombinant protein production.
Owner:GALAPAGOS NV
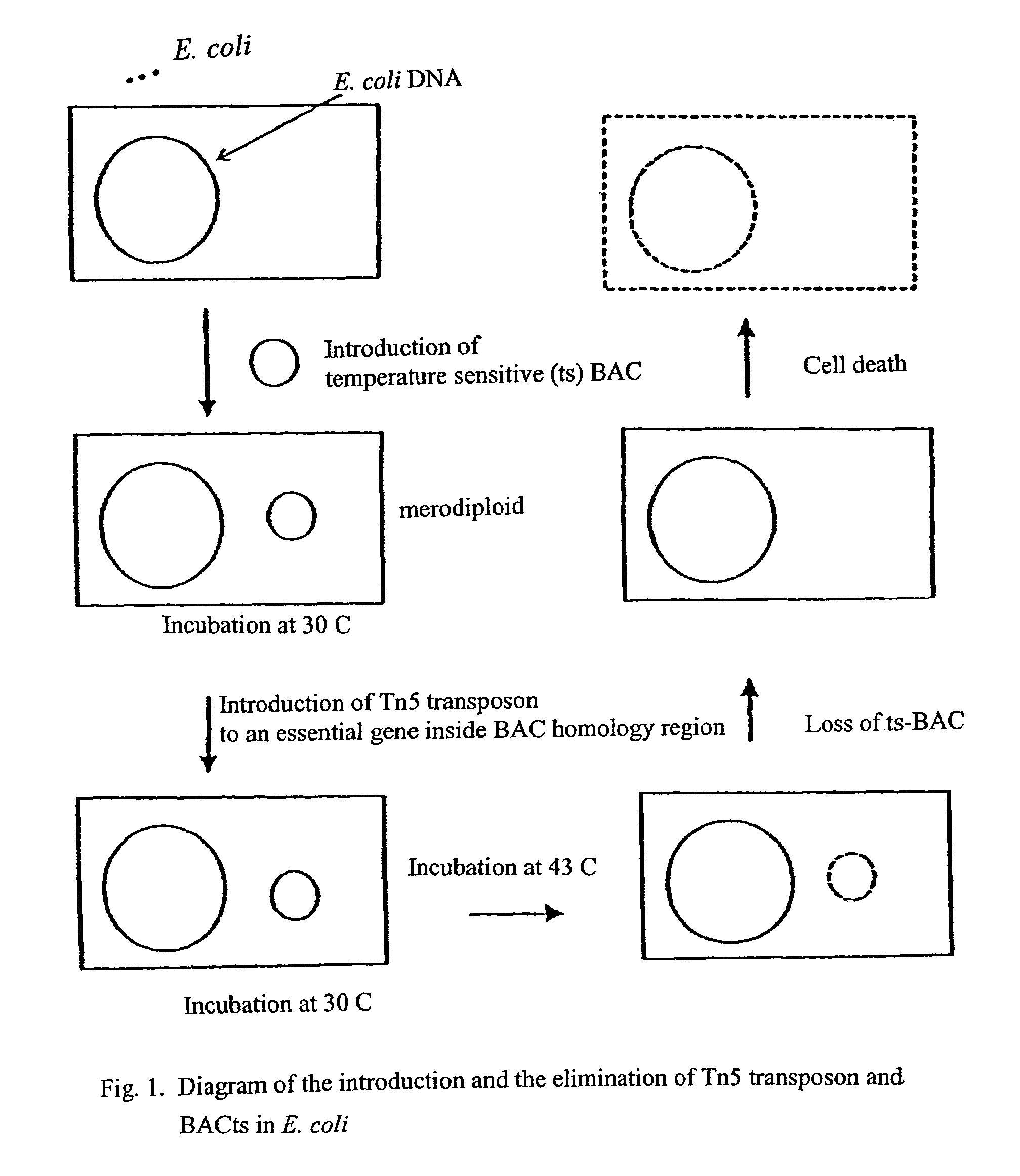
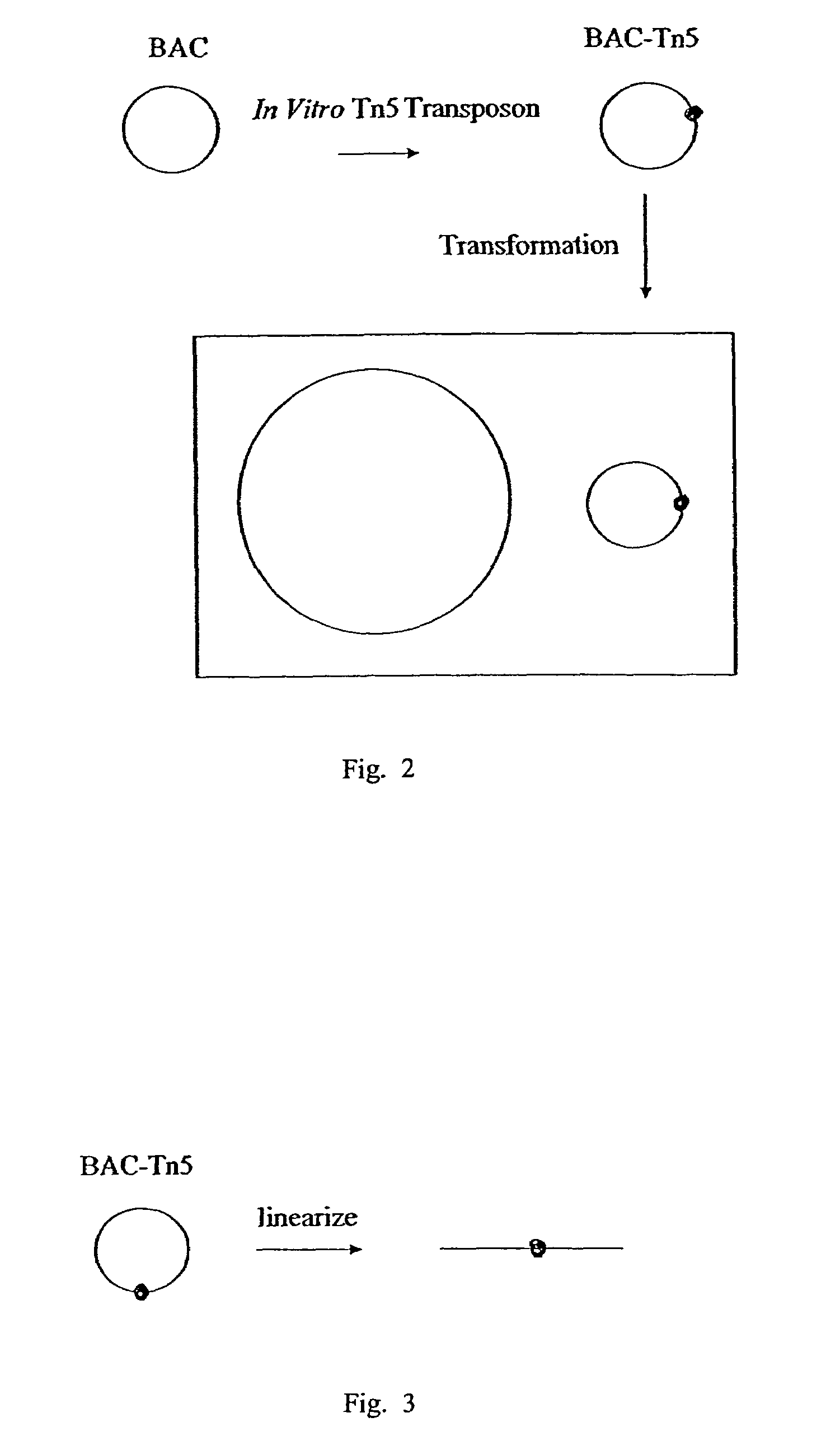
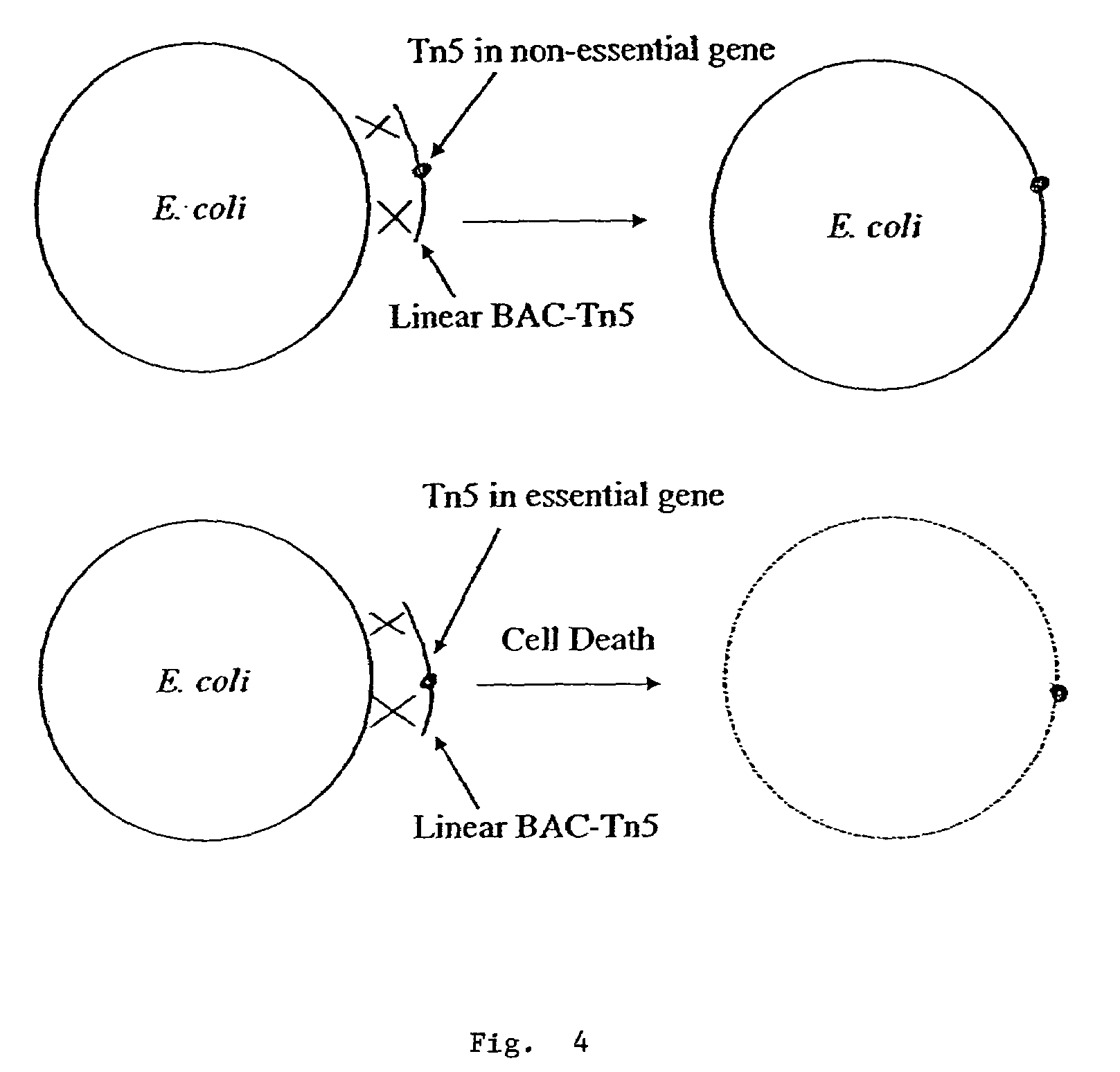
Methods for identifying an essential gene in a prokaryotic microorganism
Methods are provided for the rapid identification of essential or conditionally essential DNA segments in any species of haploid cell (one copy chromosome per cell) that is capable of being transformed by artificial means and is capable of undergoing DNA recombination. This system offers an enhanced means of identifying essential function genes in diploid pathogens, such as gram-negative and gram-positive bacteria.
Owner:CALIFORNIA INST OF TECH
Method for constructing high-flux simplified genome sequencing library
ActiveCN104694635AReduce the impact of quantitative accuracy before mixingImprove uniformityMicrobiological testing/measurementLibrary creationGenomic sequencingPhosphorylation
The invention provides a method for constructing a high-flux simplified genome sequencing library. The method comprises the following steps: digesting genome DNA of different samples by using restriction enzymes, thereby obtaining blunt-end DNA fragments with phosphorylation modification at the 5' end; adding the base A at the 3' end, connecting the DNA fragments of different samples with 5' phosphorylation and 3' cohesive ends A with joints containing different bar code sequences, connecting the products PCR, purifying and mixing the amplification products, performing low-cycle PCR amplification, thereby obtaining a target fragment amplification product of which the ratio of the joints to joint dimers is greatly reduced; and separating, purifying, and forming the high-flux simplified genome sequencing library from the purified products. The method disclosed by the invention has the advantages that the flux is high, the cost is low, and the generated library is high in sequencing quality and has very wide application prospects in researches such as high-flux SNP detection and marker development, genetic map drawing and functional gene location.
Owner:BIOMARKER TECH
Gene Expression Profile Algorithm and Test for Determining Prognosis of Prostate Cancer
ActiveUS20130196321A1Health-index calculationMicrobiological testing/measurementProstate cancerOncology
The present invention provides algorithm-based molecular assays that involve measurement of expression levels of genes, or their co-expressed genes, from a biological sample obtained from a prostate cancer patient. The genes may be grouped into functional gene subsets for calculating a quantitative score useful to predict a likelihood of a clinical outcome for a prostate cancer patient.
Owner:MDXHEALTH
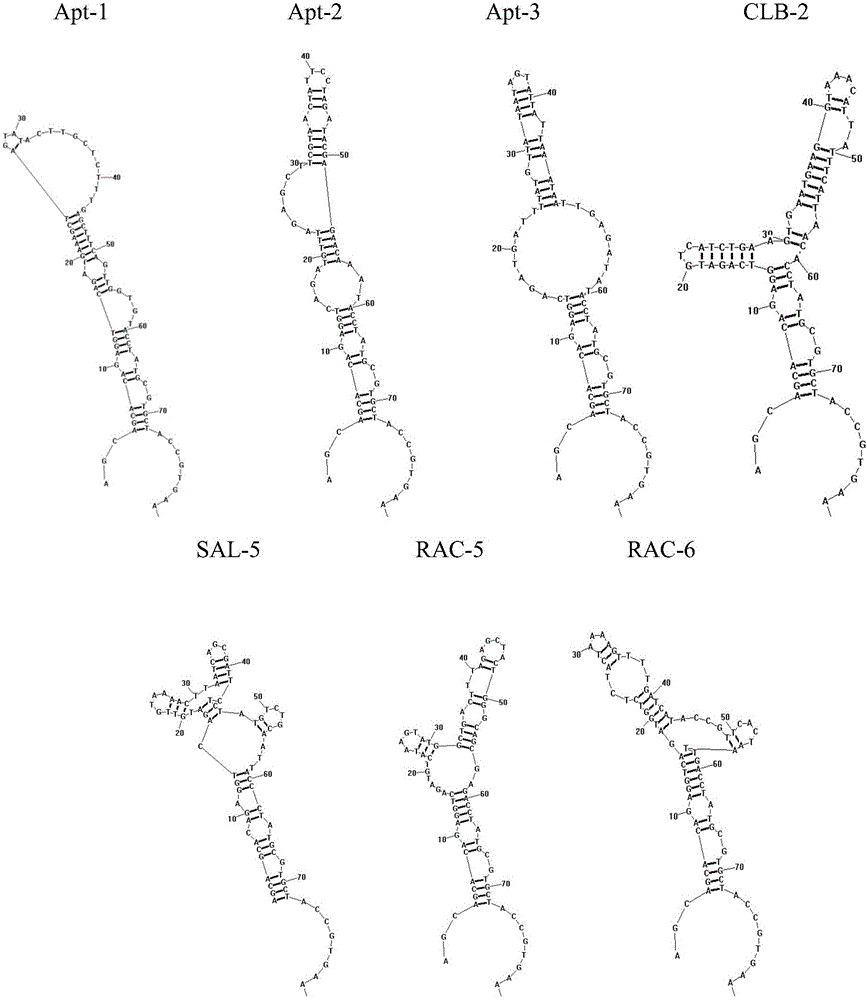
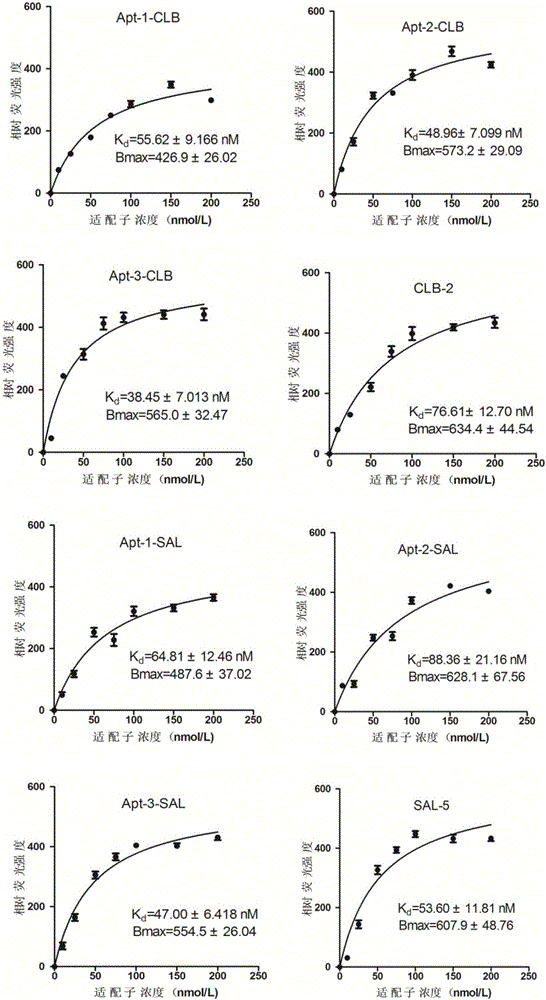
A group of oligonucleotide aptamers for identifying clenbuterol hydrochloride, salbutamol and ractopamine with high specificity
ActiveCN105886512AEasy accessAccurate acquisitionBiological testingDNA/RNA fragmentationAptamerFluorescence
The invention provides a group of oligonucleotide aptamers Apt-1, Apt-2 and Apt-3 which are capable of simultaneously identifying clenbuterol hydrochloride and salbutamol, an oligonucleotide aptamer CLB-2 which is capable of identifying the clenbuterol hydrochloride with high specificity, an oligonucleotide aptamer SAL-5 which is capable of identifying the salbutamol with high specificity and two oligonucleotide aptamers RAC-5 and RAC-6 which are capable of identifying ractopamine with high specificity. Through an SELEX (Systematic Evolution of Ligands by Exponential Enrichment) technology based on Fe3O4 magnetic nanoparticle separation, a random oligonucleotide library is immobilized on avidin-enveloped magnetic nanoparticles by virtue of a complementary chain of a biotinylation marker, and the oligonucleotide aptamers, which are high in specific affinity, are finally obtained by conducting screening by 16 turns. The aptamers are broad in application prospect; and the aptamers, by virtue of marker function genes or fluorescent dyes, are applicable to detection of the clenbuterol hydrochloride, the salbutamol and the ractopamine in food; therefore, a new choice is provided for existing detection methods which depend on antibodies.
Owner:JIANGNAN UNIV
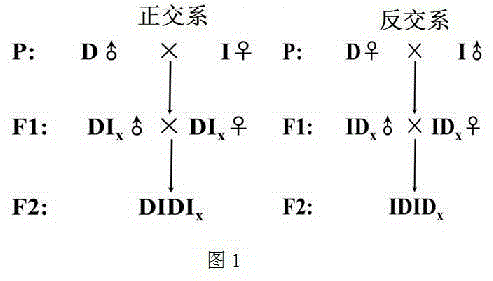
Establishment method of goat resource population
InactiveCN105123621AImprove economyImprove international competitivenessAnimal husbandryNational levelReciprocal cross
The invention belongs to the field of establishment of a population genetic resource bank, and provides a scientific establishment method of a goat resource population bank, and provides a goat reference family system establishment case, and relates to two national-level genetic resource varieties, namely, a Dazu black goat and an Inner Mongolia Cashmere goat (Alxa type). According to the resource population establishment method, forward cross and reciprocal cross of Dazu black goat and Inner Mongolia Cashmere goat (Alxa type) are utilized, random mating of genetic relationship is avoided for F1, so that Mendel genetic characters are separated, a goat economic character QTL or a functional gene is located by virtue of a genome means, and a relatively-precise genetic mark is provided for the goat breeding. The genetic resource bank comprises goat population, a genealogy inforamtion database, a phenotypic character database, a DNA sample library, an RNA sample library and a genetic variation information database.
Owner:SOUTHWEST UNIVERSITY
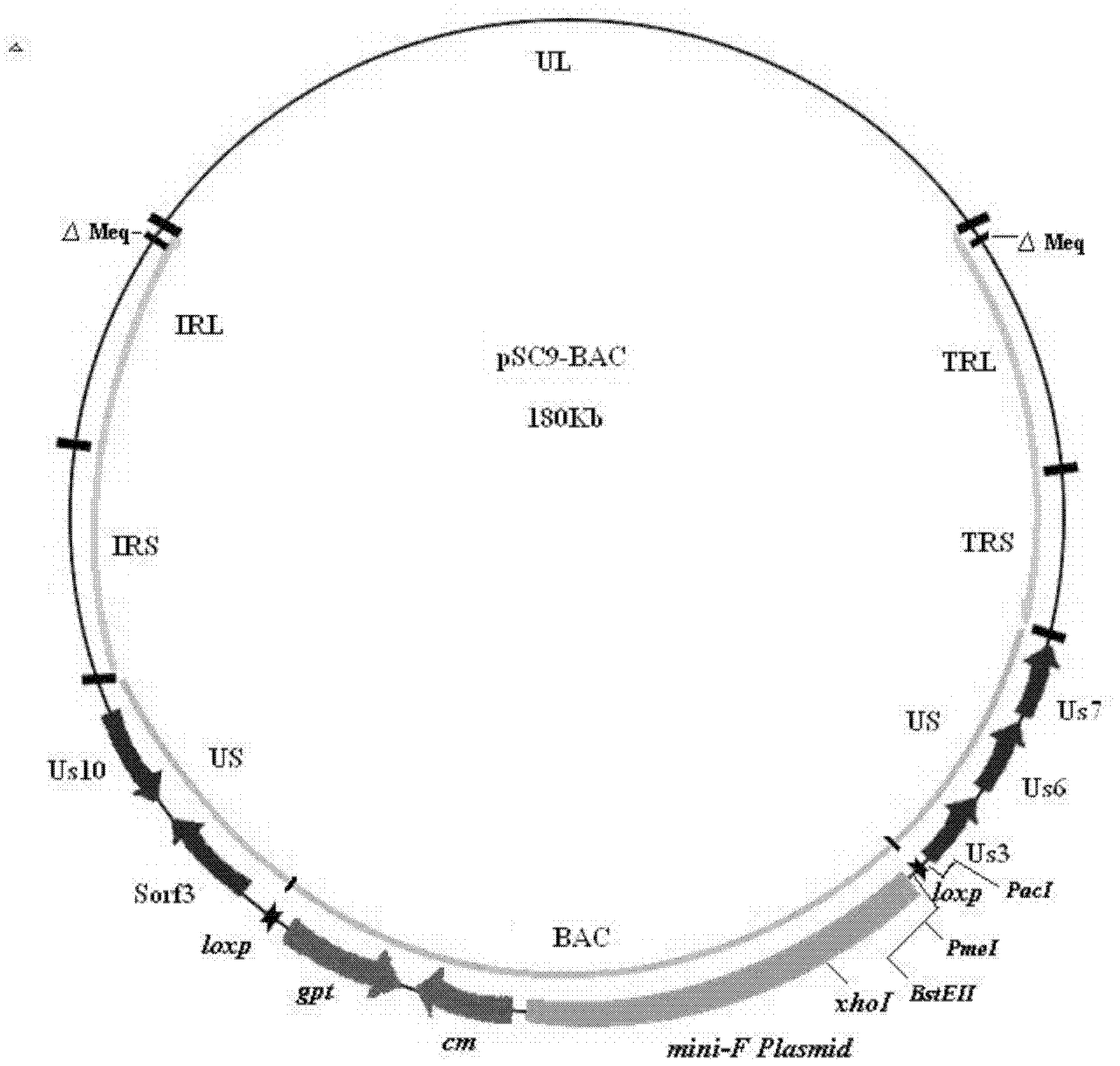
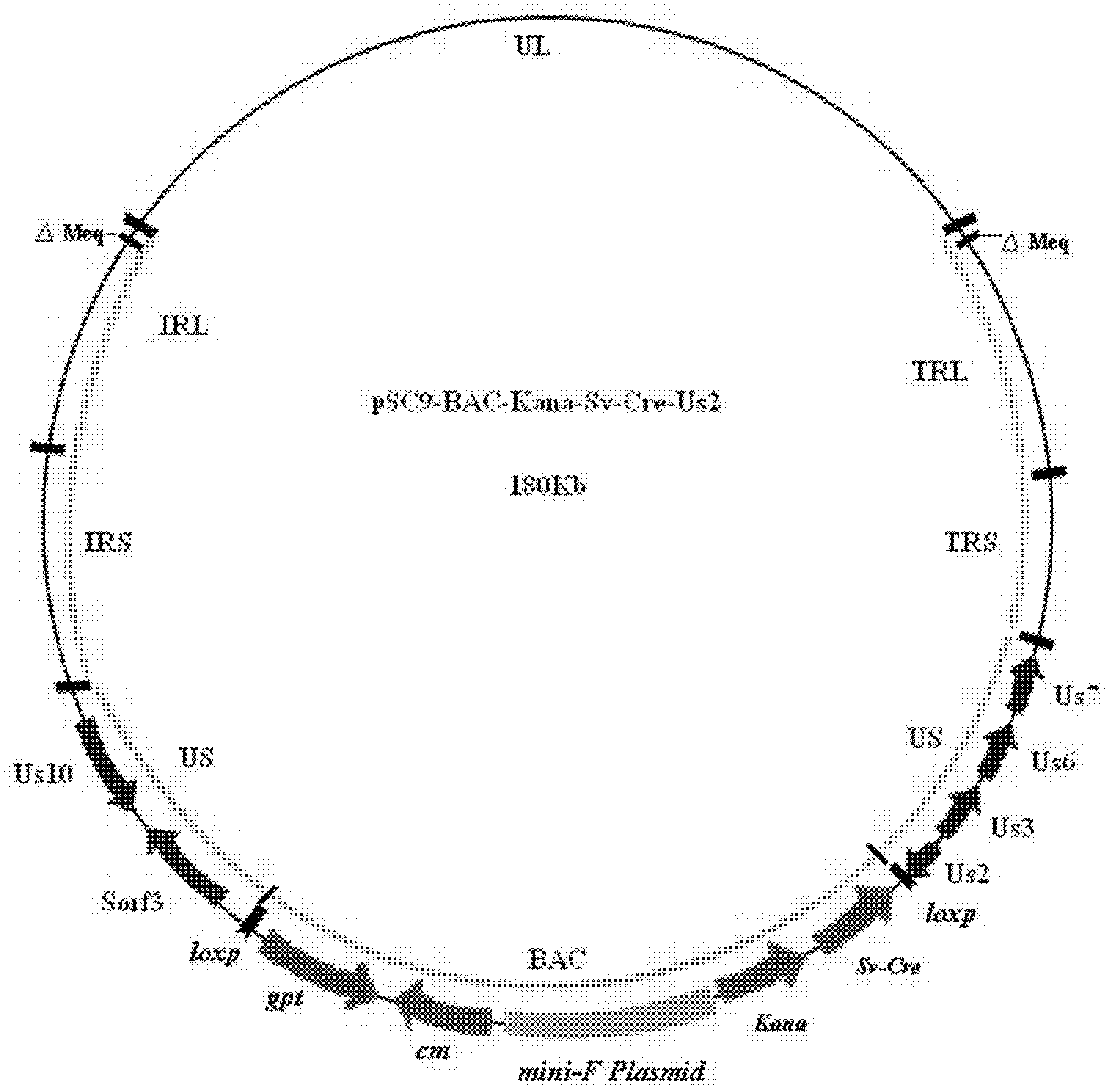
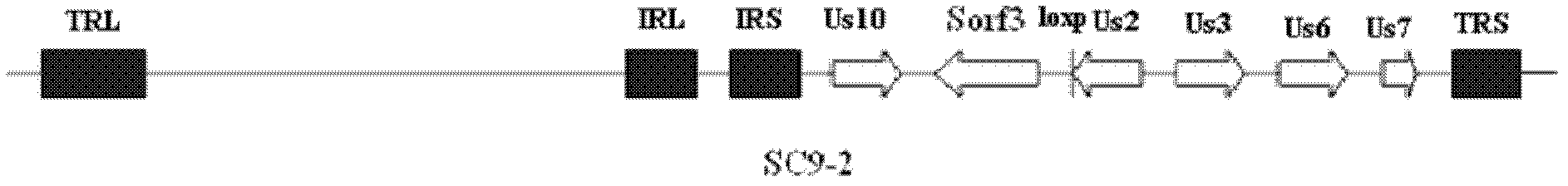
Construction and application of recombinant Chicken Marek's Disease Virus SC9-1 strain and SC9-2 strain
The invention relates to a construction and an application of recombinant Chicken Marek's Disease Virus SC9-1 strain and SC9-2 strain. The recombinant virus construction method solves the technical problem that kanr gene cannot be knocked out again by a present method after kanr gene containing flp recognition sites at two ends is continuously used twice to knock out two specific functional genes on the same virus genome. The obtained recombinant virus MDV SC 9-1 strain and MDV SC 9-2 strain are used as production strains of Marek's Disease live vaccine. The prepared vaccine prevents the very virulent or emerging very virulent plus MDV induced chicken Marek's disease. The protective immunity effect of the vaccine is superior to CVI988 / Rispens strain vaccine which is mostly widely used in foreign and domestic markets at present. The antigenicity of the recombinant virus is more similar to that of Chinese epidemic strain than the antigenicity of the similar virus rMd5deltameq which has been published in the United State. The strains provided by the invention will not induce tumor and has no immune suppression effect. Therefore, the strains are more applicable to China.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Recombined streptomyces nodosus capable of producing amphotericin B and application thereof
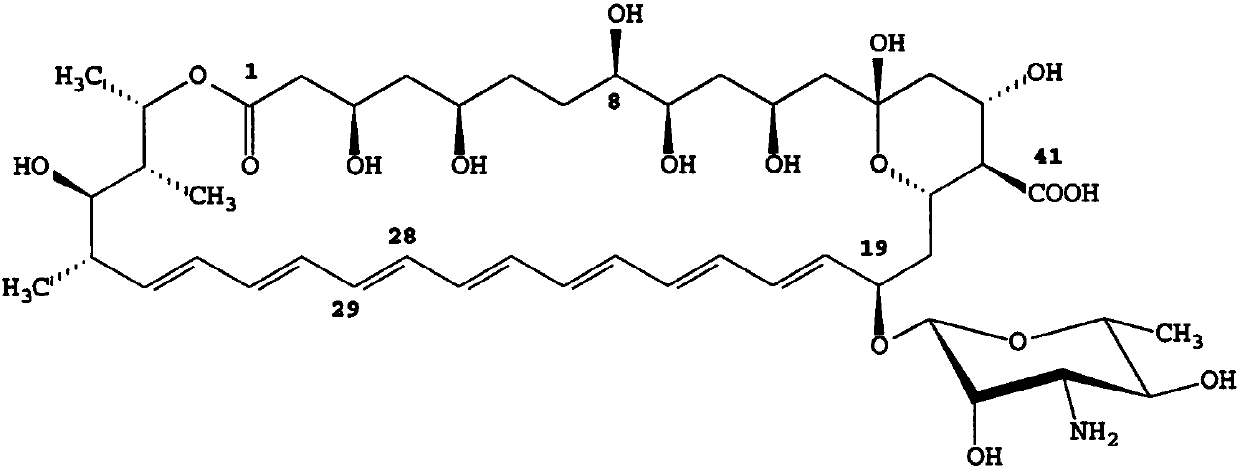
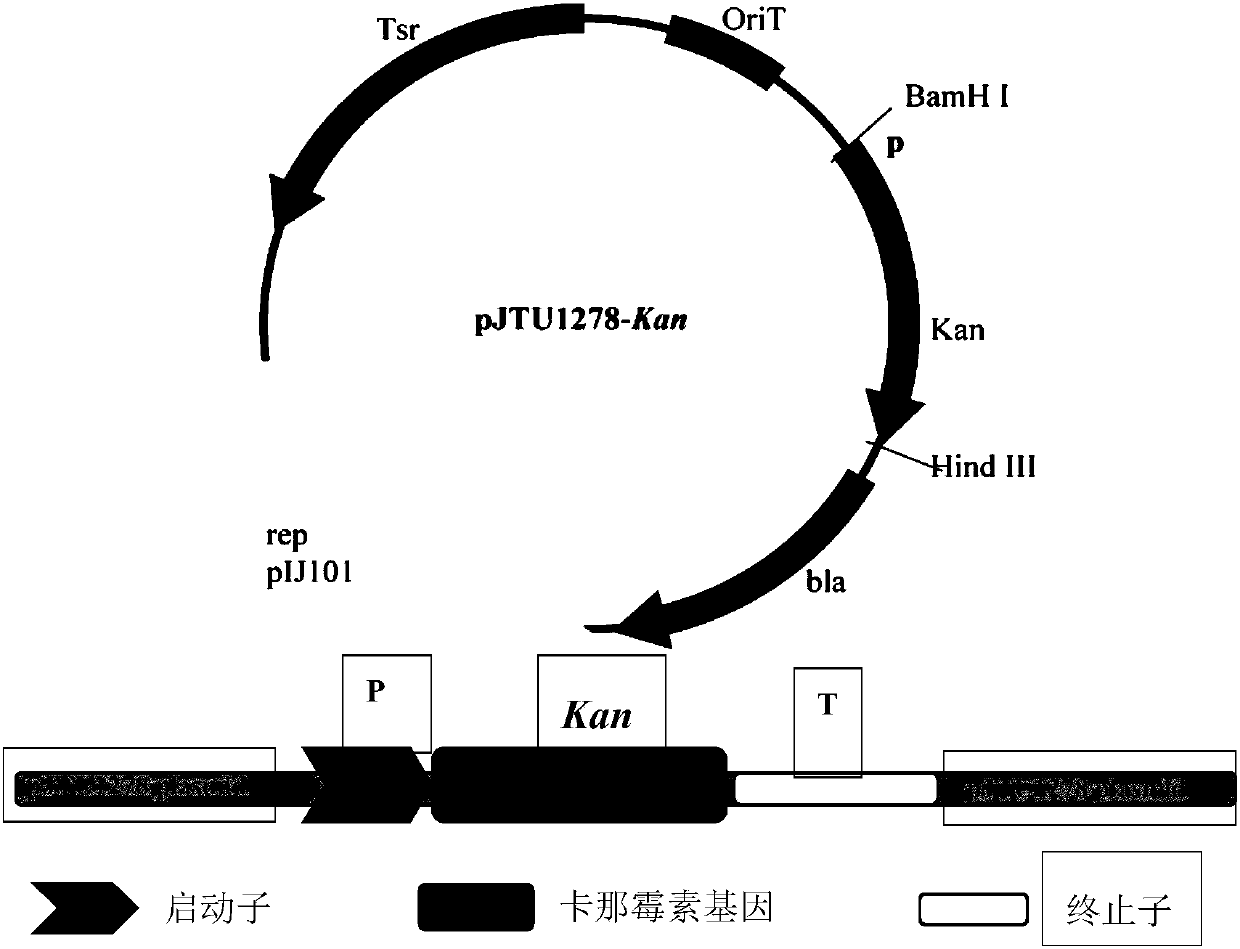
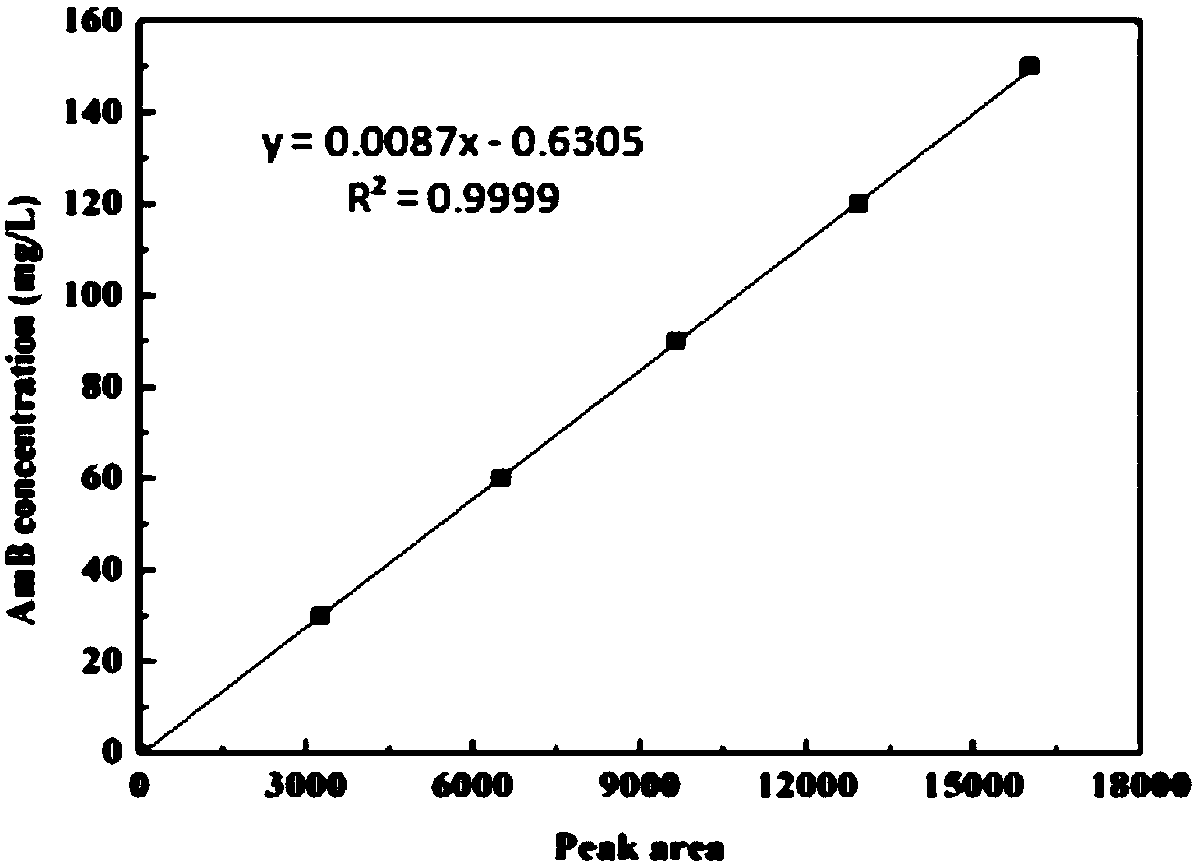
ActiveCN107893048AHigh purityQuality improvementBacteriaTransferasesBiotechnologyS-Adenosyl-l-methionine
The invention discloses recombined streptomyces nodosus capable of producing amphotericin B and application thereof. The recombined streptomyces nodosus is obtained by introducing a kanamycin resistance gene sequence showed in SEQ ID NO.1 into streptomyces nodosus ZJB2016050; by introducing the kanamycin resistance gene, produced strains which have no resistance previously have kanamycin resistance, the purity and quality of the strains of seed culture mediums are improved, and strain contamination phenomena can be prevented from occurring during seed cultivation in a lab. By expressing a function gene, namely vitreoscilla hemoglobin (vhb), the yield of AmB is increased by 15%; by expressing a function gene, namely the S- adenosylmethionine synthetase gene (metk), the yield of AmB is increased by about 40%.
Owner:ZHEJIANG UNIV OF TECH
Novel capture and enrichment technology for targeting nucleic acid molecules
InactiveCN103602658AHigh detection sensitivityIncreased chance of collisionMicrobiological testing/measurementDNA preparationHuman DNA sequencingNucleic acid molecule
The invention discloses a novel capture and enrichment technology for targeting nucleic acid molecules. The targeting nucleic acid molecule fragments are hybridized through special capture probes; and probe molecules marked with acrylamide groups are fixed in acrylamide glue, so that the targeting nucleic acid molecule fragments are captured. The technology can be used for functional gene capture and high-throughput sequencing analysis of microbial community, fishing and high-throughput sequencing analysis of genes related to diseases in human genome, and fishing and analysis of signature sequences of environment pathogenic microorganisms. Besides, the technology can be used for development of diagnostic kit for a plurality of molecular diseases and pathogenic microorganisms. The technology is relatively low in cost. Glue efficiency of acrylamide molecules is far higher than connection efficiency of magnetic beads and probes, so that capture efficiency is greatly increased.
Owner:SOUTHEAST UNIV
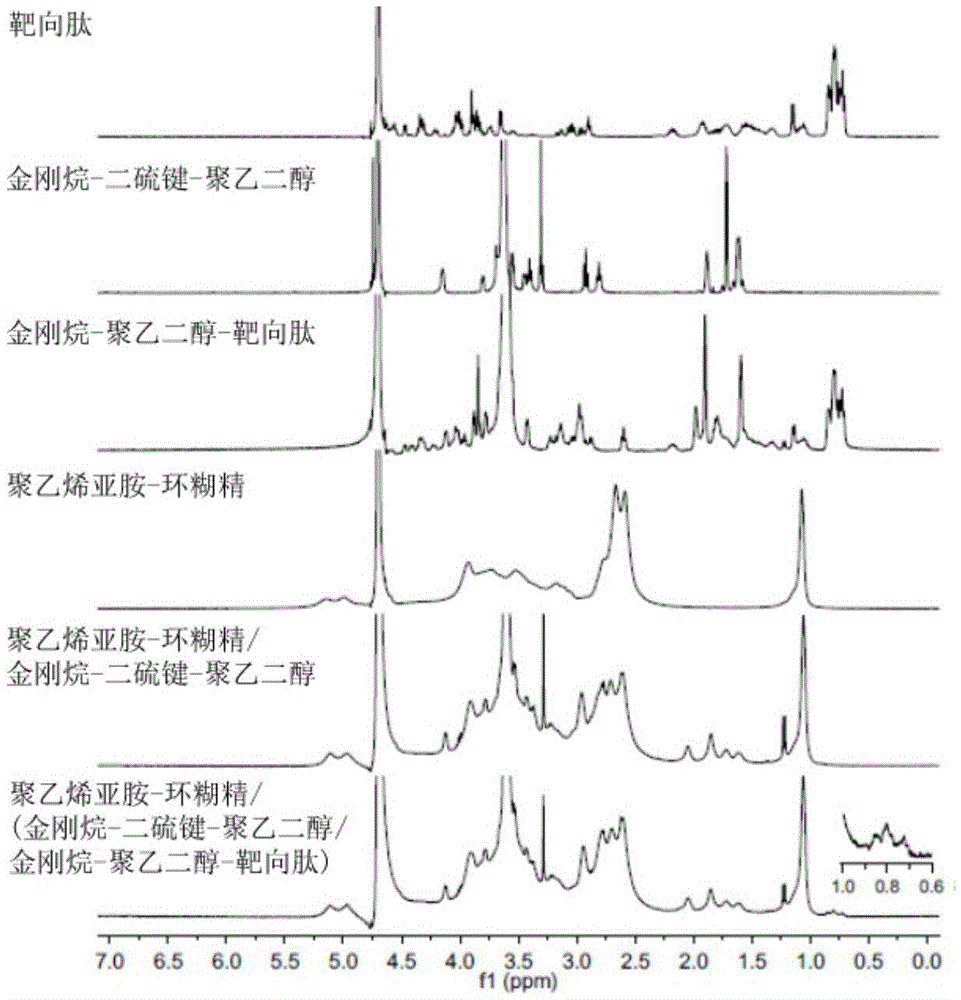
Multifunctional supramolecular gene delivery system based on polyethyleneimine-cyclodextrin and preparation method thereof
InactiveCN105463024AStrong transformationDegraded fromOther foreign material introduction processesGene deliveryPolyethylene glycol
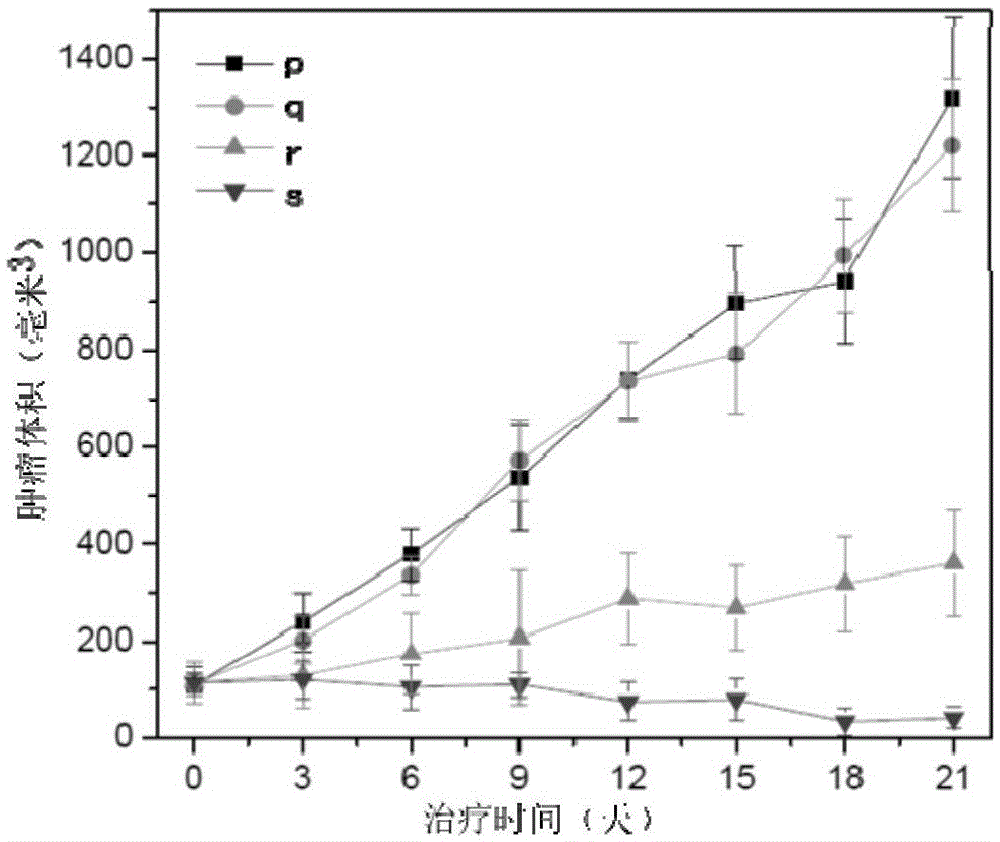
The invention discloses a multifunctional supramolecular gene delivery system based on polyethyleneimine-cyclodextrin and a preparation method thereof. A polyethyleneimine-cyclodextrin polycationic compound serves as a framework, adamantane-disulfide bond-polyethylene glycol having oxidation reduction reactivity and adamantane-polyethylene glycol-targeting peptide having targeting performance serve as modified groups, polyethyleneimine-cyclodextrin is combined with adamantane-disulfide bond-polyethylene glycol and adamantane-polyethylene glycol-targeting peptide through a self-assembling principle, and a subjective body and an objective body act to form a supramolecular gene carrier. The delivery system is multifunctional, has oxidation reduction sensitivity, specificity for targeting liver cancer and excellent gene transfection efficiency, and can carry functional genes to efficiently treat liver malignant tumor.
Owner:ZHEJIANG UNIV
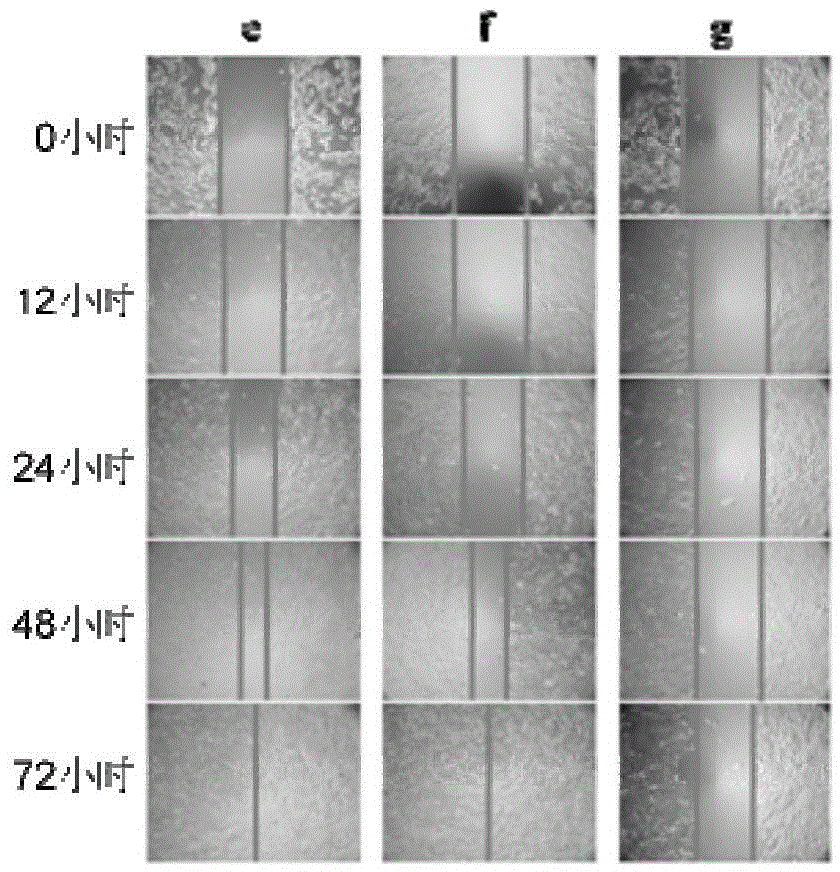
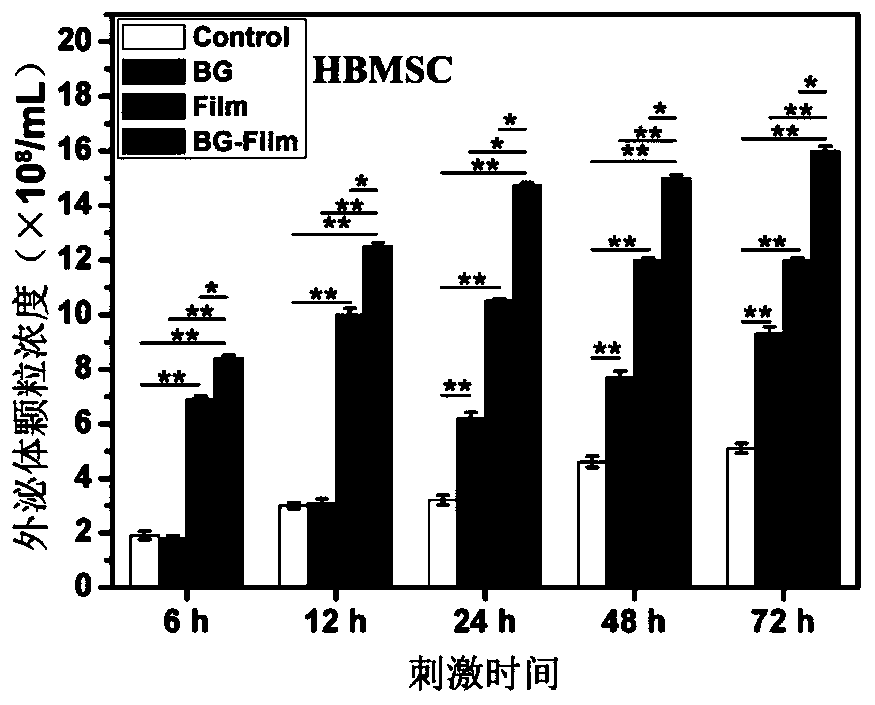
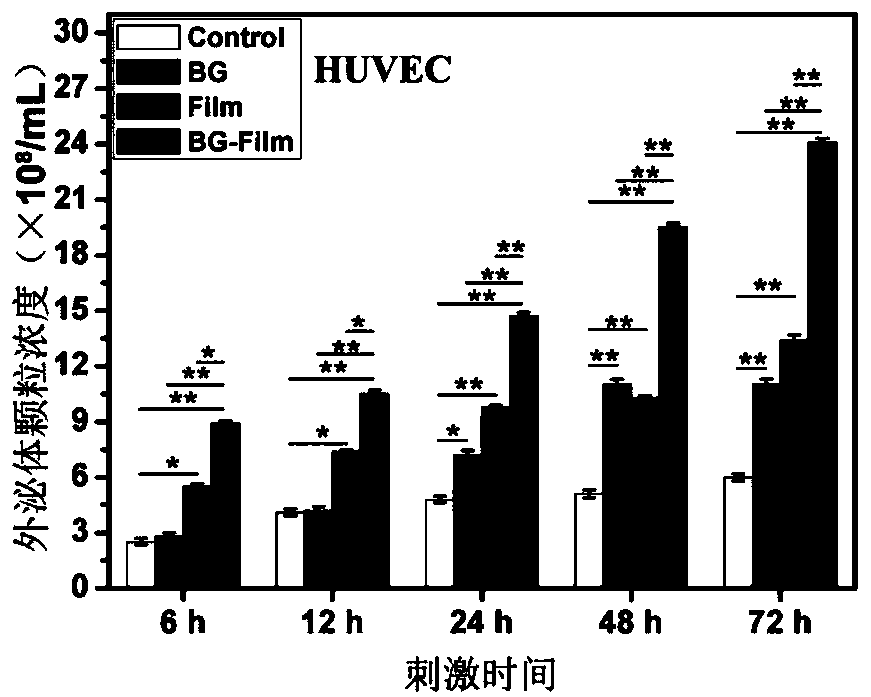
Method and application for promoting cells to secrete exosomes
ActiveCN110129266ARapid functional differentiationIncrease productionSkeletal/connective tissue cellsCell culture supports/coatingFiberTissue repair
The invention discloses a method and application for promoting cells to secrete exosomes, and relates to the fields of biological materials, tissue engineering and regenerative medicines. According tothe method, the cells are stimulated by active signals of a biological material, and the exosomes generated by the cells are collected, and are used for tissue wound treatment. The operation route ofthe method and the application scheme of the obtained exosomes are described in detail, under the stimulation effects of bioactive glass chemical signals, electrostatic spinning fiber membrane structure signals and chemical / structural combination signals, more exosome particles can be secreted by mesenchymal stem cells and umbilical vein endothelial cells, and the exosomes can promote the corresponding target cells to express higher levels of functional genes, secrete more multifunctional proteins and carry out deep functional differentiation, so that the tissue wound surface can be rapidly repaired. The exosomes obtained by the method disclosed by the invention have the characteristics of higher yield and stronger tissue repair promotion function.
Owner:SHANGHAI JIAO TONG UNIV
High-throughput cell transfection device and methods of using thereof
ActiveUS7687267B2Microbiological testing/measurementOther foreign material introduction processesFunctional analysisThroughput
Transfecting biology cells with nucleic acid molecules (DNA, siRNA) is an essential prerequisite in elucidating how genes function in complex cellular context and how their activities could be modulated for therapeutic intervention. Traditionally studies are carried out on a low throughput gene-by-gene scale, which has created a huge bottleneck in functional genomic study and drug discovery. Development of high-throughput cell transfection technology will permit functional analysis of massive number of genes and how their activities could be modulated by chemical or biological entities inside cells. This invention describes design, construction of device and apparatus for high throughput effective cell transfection. Procedures and protocols for using the device and apparatus are also described in the application. Novel methods of using the device in cell-based assays are also disclosed.
Owner:RATIONAL BIOTECH
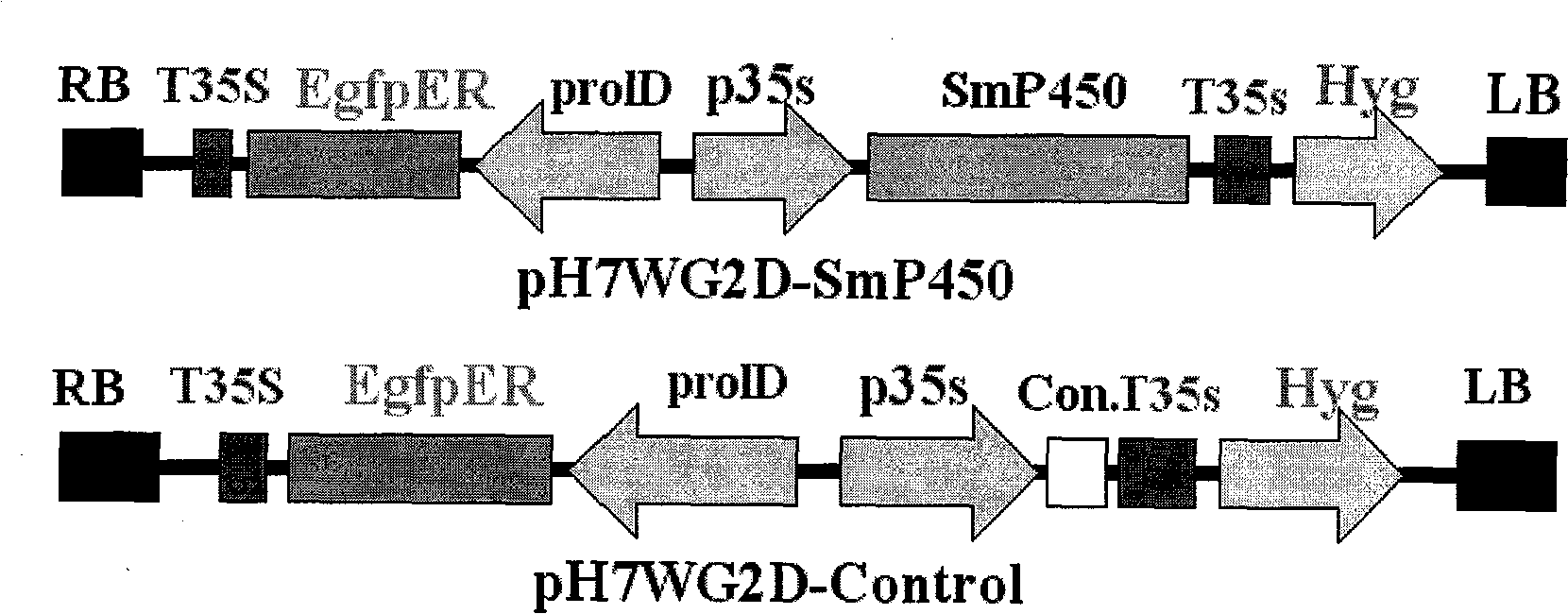
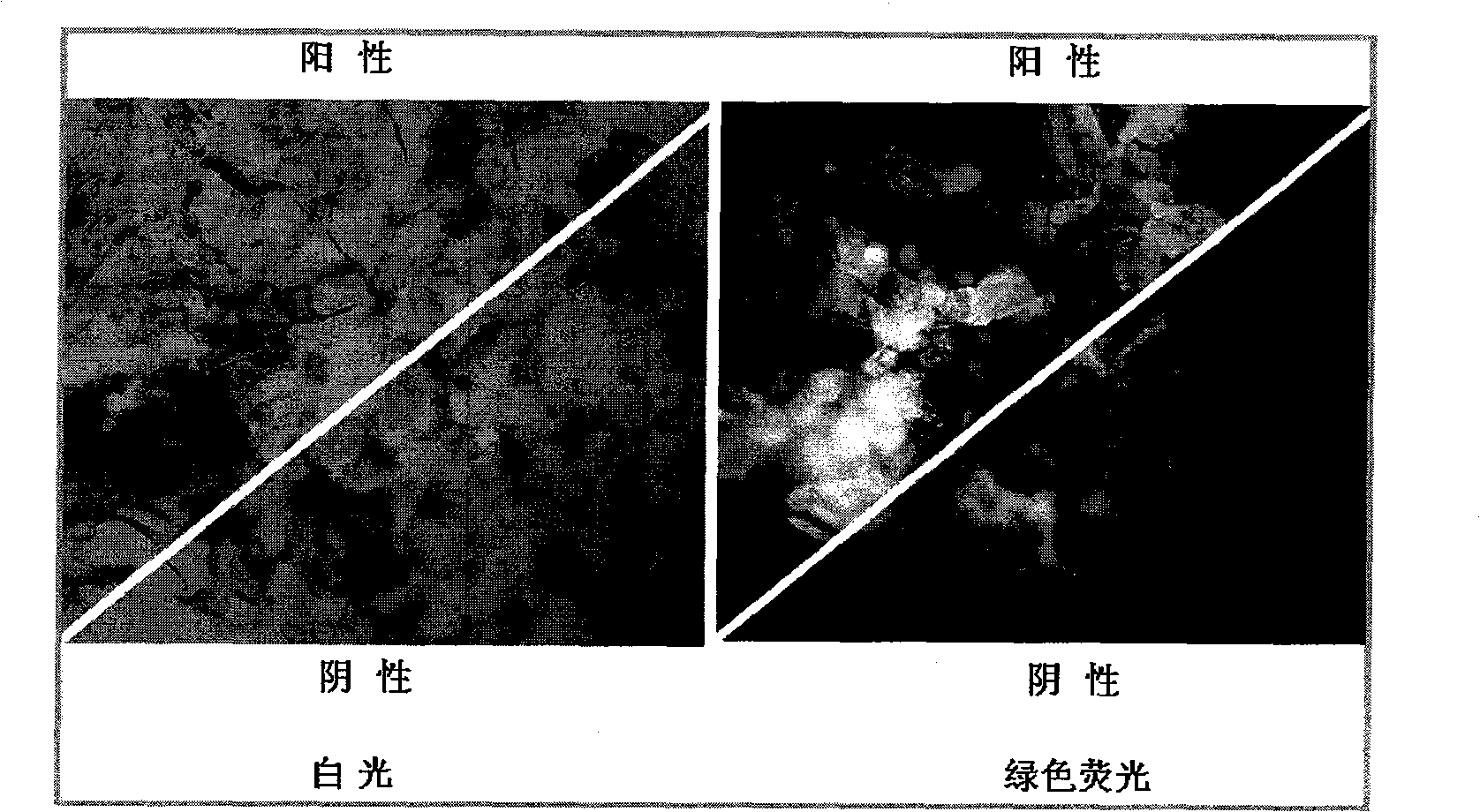
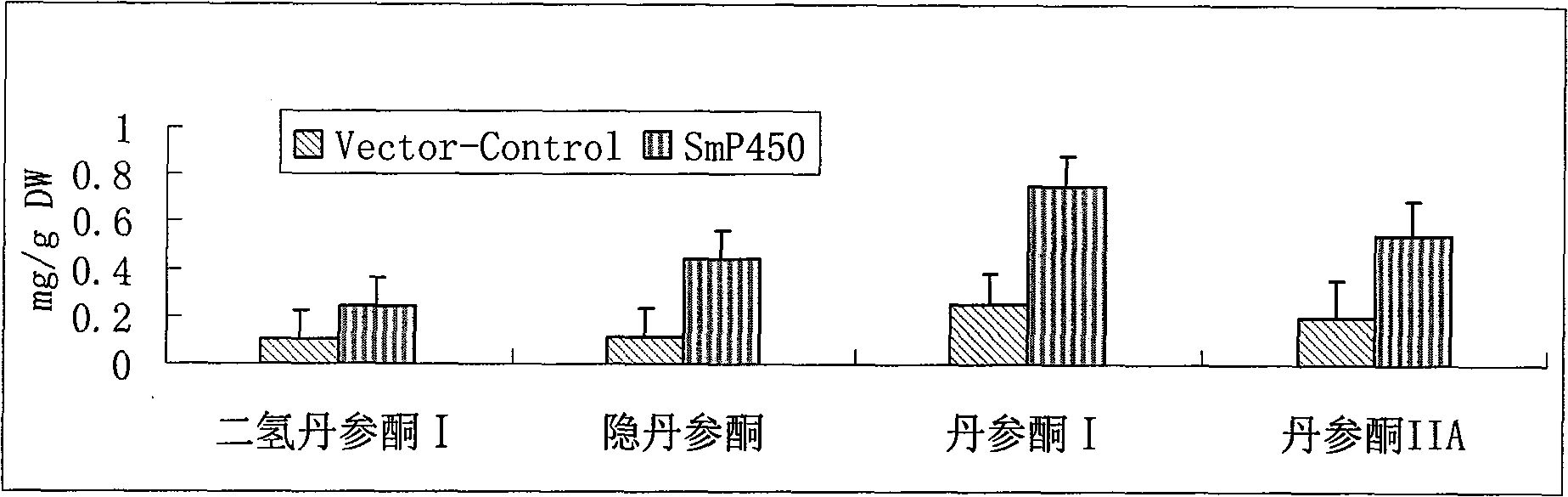
Salviamiltiorrhizabge-cytochrome P450 (SmP450) gene as well as coded protein and application thereof
InactiveCN101928715AIncrease contentRelieve severe plaque deficiencyOxidoreductasesFermentationSalvia miltiorrhizaNucleotide
The invention discloses a cytochrome P450 (SmP450) gene as well as a coded protease and application thereof. The gene is obtained by clone from salviamiltiorrhizabge by utilizing a cDNA chip technology, which fills the blank of separating and cloning a structure-modification type functional gene which is cytochrome P450 (SmP450) gene from a rare traditional Chinese medicine of salviamiltiorrhizabge. The cytochrome P450 (SmP450) gene has a nucleotide sequence shown as SEQ ID NO.1, or a homological sequence adding, substituting, inserting or lacking one or more nucleotides or allelic genes and derived nucleotide sequence thereof. The protein coded by the cytochrome P450 (SmP450) gene has an amino acid sequence shown as SEQ ID NO.2 or a homological sequence adding, substituting, inserting or lacking one or more amino acids. The cytochrome P450 (SmP450) can improve the content of tanshinone which is diterpene active element in the salviamiltiorrhizabge by the biotechnology, is beneficial to the quality improvement of the salviamiltiorrhizabge, can be used for the breeding selection and has good application prospect.
Owner:INST OF CHINESE MATERIA MEDICA CHINA ACAD OF CHINESE MEDICAL SCI
Method for converting disease-resistance genes of rice and obtaining transgenic descendants without selective markers
InactiveCN102703499AQuick screeningReduce workloadMicrobiological testing/measurementPlant tissue cultureDiseaseFluorescence
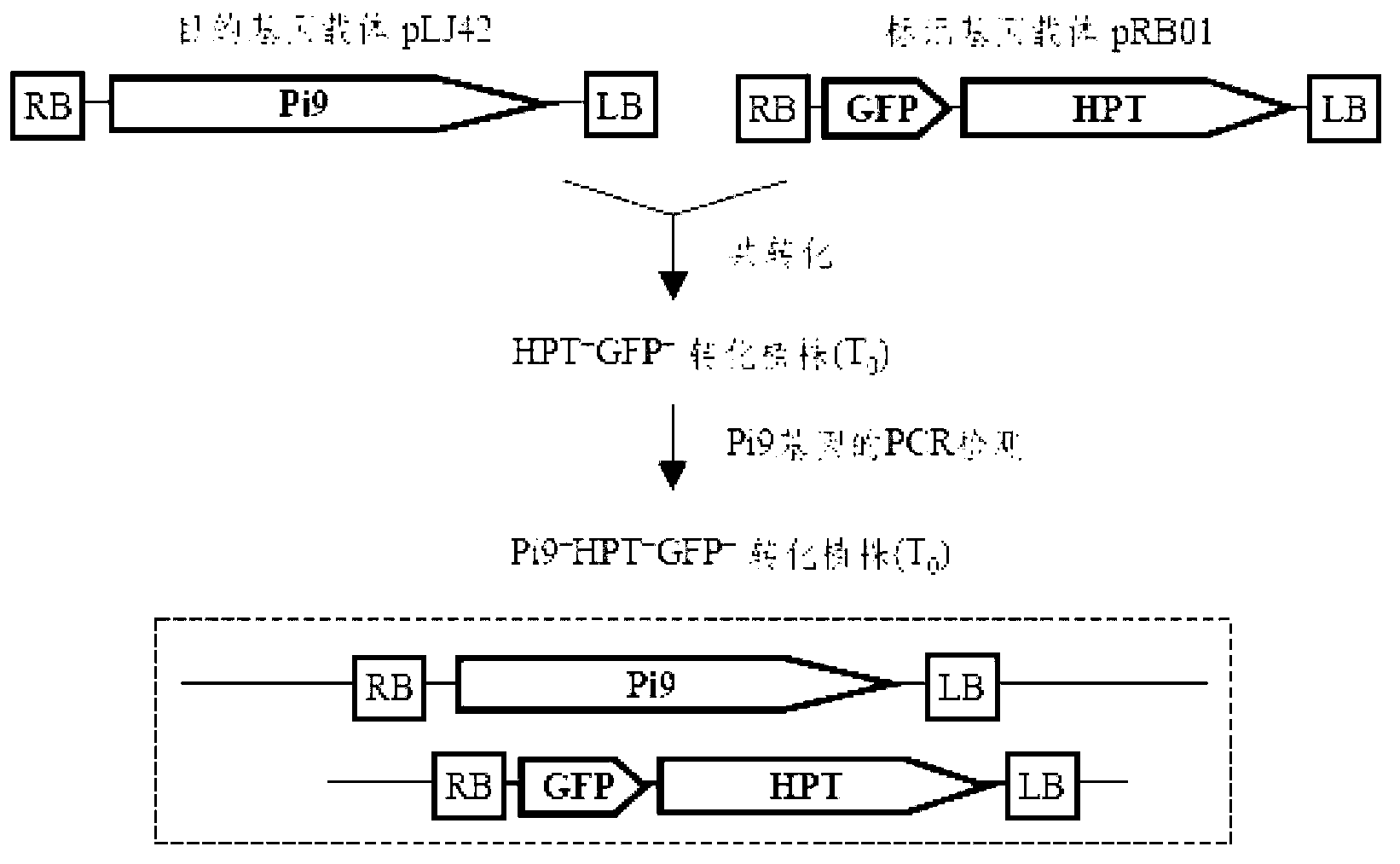
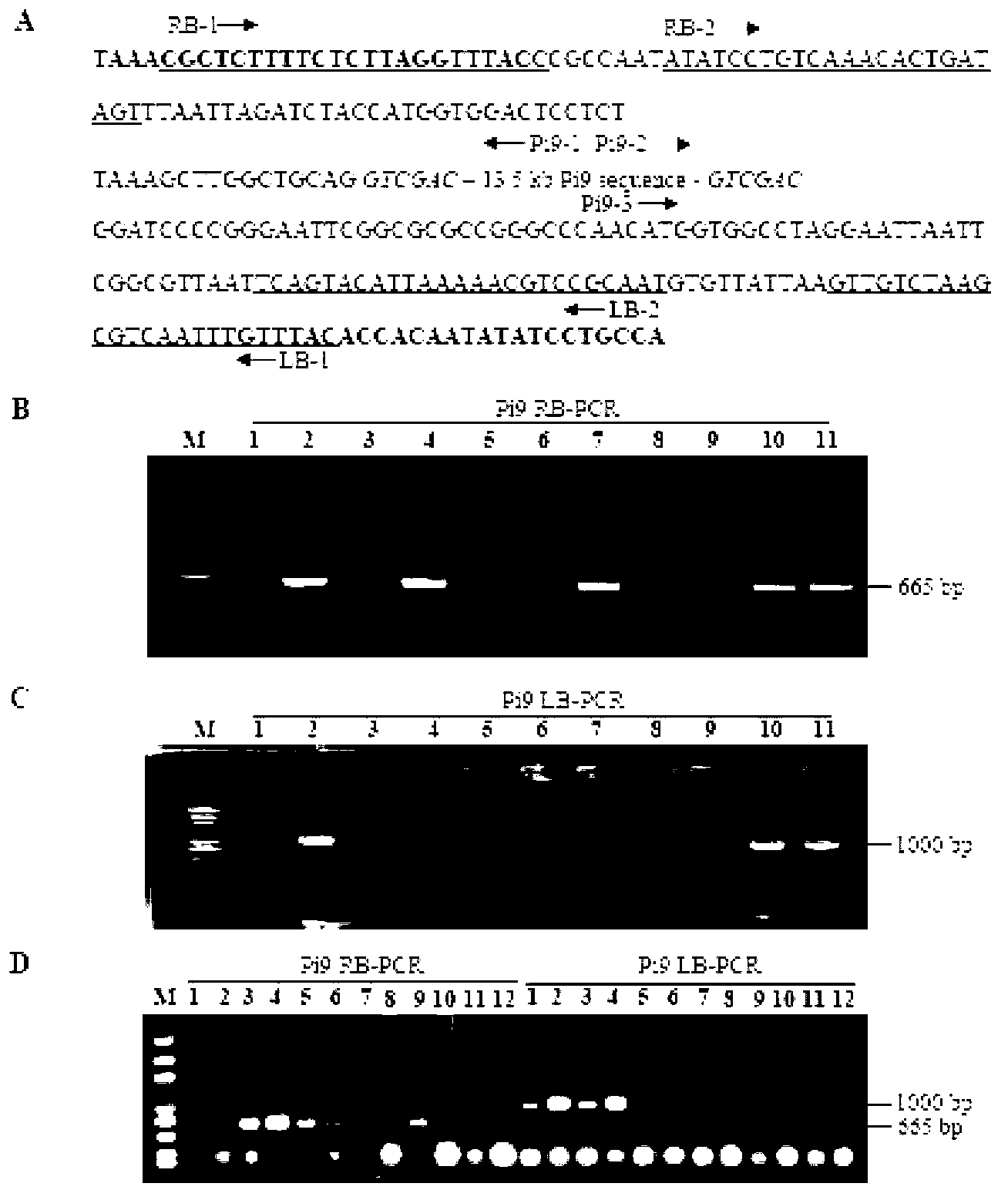
The invention relates to a method for converting disease-resistance genes of rice and obtaining transgenic descendants without selective markers. The method comprises the following steps: first, cloning green fluorescent protein (GFP) and hygromycin phosphotransferase (HPT) on a marker gene carrier, placing a target gene in other T-DNA carrier, mixing strains of agrobacterium tumefaciens carrying the marker gene carrier and strains of agrobacterium tumefaciens carrying the target gene carrier and converting callus of the rice, performing PCR (polymerase chain reaction) detection on the disease-resistance genes of the target gene carrier by a specific primer, and screening to obtain co-transformation plants (T0); then, screening the marker gene plants with positive GFP rapidly and massively by means of a desk lamp fluorescence detector in segregative generations (T1 or T2) of the co-transformation plant for removing, and performing PCR detection for disease-resistance genes to the plants with negative GFP so as to obtain the individuals without selective markers of the transgenic disease-resistance genes. The method can be applied to transgenic breeding without selective markers of rice blast-resistant genes or other functional genes of the rice, and enhances the disease resistance of the rice or improves other agronomic traits.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
Flavoprotein oxidase genes of macleaya cordata in synthesis of sanguinarine and chelerythrine and application of flavoprotein oxidase genes
ActiveCN106047904AAchieve synthesisAchieve in vitro synthesisOxidoreductasesGenetic engineeringHeterologousFlavoprotein
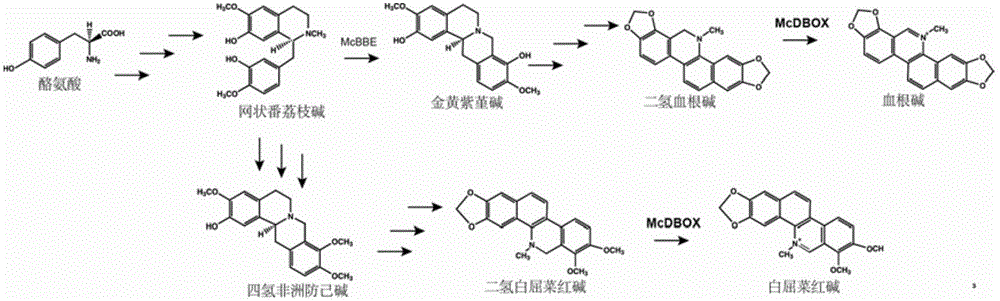
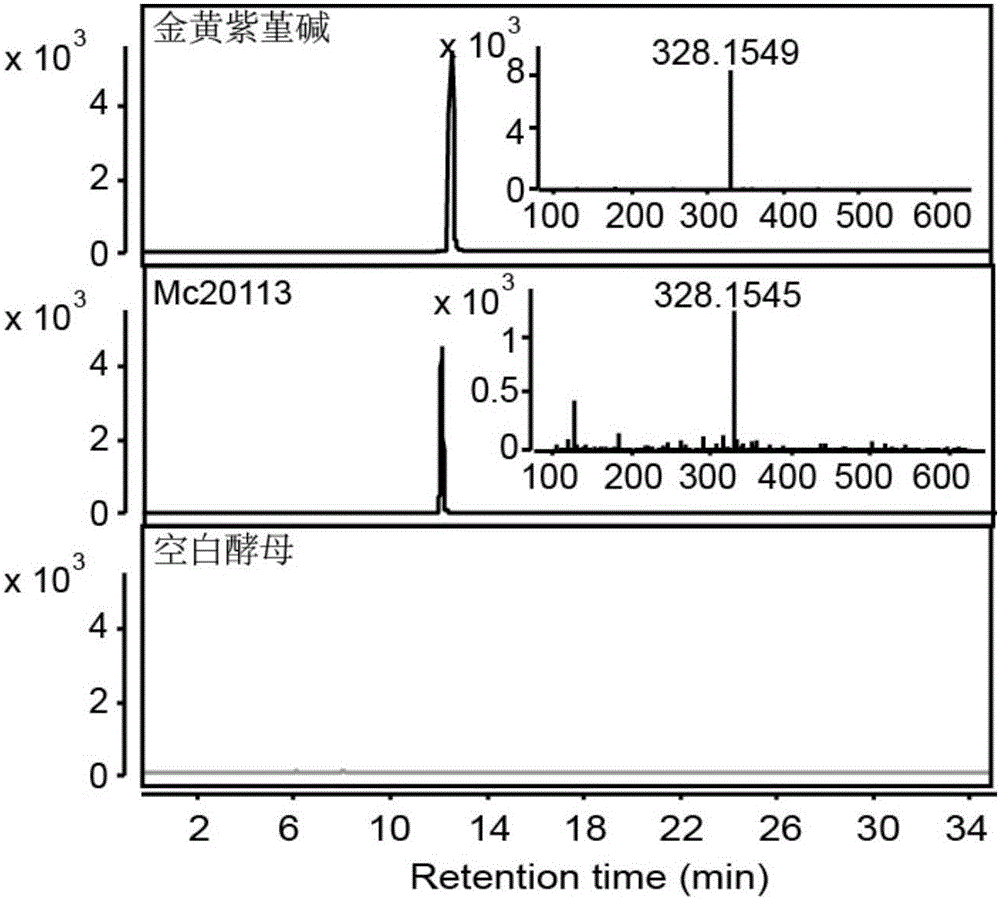
The invention discloses flavoprotein oxidase genes of macleaya cordata in synthesis of sanguinarine and chelerythrine and application of the flavoprotein oxidase genes. Three flavoprotein oxidase genes joining in synthesis of sanguinarine and chelerythrine are found in a macleaya cordata genome for the first time, including genes Mc20113, Mc6407 and Mc6408. Steps of reaction are respectively verified by using a brewer's yeast system through upstream precursor feeding, and synthesis of sanguinarine and chelerythrine and midbodies can be achieved. The conversion efficiency of same functional genes of macleaya cordata and opium poppy is further compared by using a brewer's yeast heterologous expression system, and the result shows that the conversion rate of the gene of macleaya cordata is remarkably higher than that of opium poppy. The invention further discloses a molecular mechanism of synthesis of sanguinarine in macleaya cordata, and a theoretic basis and molecular assistant breeding targets are provided for breeding of macleaya cordata with high contents of sanguinarine and chelerythrine; and meanwhile, precious experience is provided for in-vitro artificial synthesis of sanguinarine and chelerythrine.
Owner:MICOLTA BIORESOURCE INC LTD
Trace and complex DNA amplification method
The present invention relates to a high sensitivity, high fidelity, high uniformity and high efficiency amplification technology of trace and complex DNA. The technology system is constant temperature DNA amplification optimized by high concentration trehalose and guided by DNA polymerase, wherein balanced and efficient amplification of DNA in different regions of the whole genome or a DNA library sequence can be maintained in the context of effective inhibition on non-specific amplification by the amplification system. The method has the following characteristics that: sensitivity is high; balanced amplification of different region sequences can be maintained, and the amplification product has high fidelity (minimum deviation) and high uniformity; and the total reaction volume has high flexibility, and can be conveniently adjusted according to requirements. In addition, the technology provides broad application prospects in single cell level and trace genome DNA amplification and gene detection, pretreated function genome associated DNA library (spectrum) analysis, and other fields.
Owner:SHANGHAI GREENGLOBE BIOTECH
Detecting protein of transcriptional factor through probe of digestion FRET marke nucleic acid of nuclease
InactiveCN1727895AEfficient analysisEfficient comprehensive analysis and detectionMicrobiological testing/measurementChemiluminescene/bioluminescenceRegulation of gene expressionNucleic Acid Probes
A method for detecting transcription factor protein by nucleic acid probe with nuclease protection PRET label includes preparing fluorescence labeled nucleic acid probe, carrying out combination reaction of nucleic acid probe with transcription factor protein, adding nuclease in combination reaction system to carry out enzyme nick reaction and detecting reaction system by fluorescence to reflect expression and activation level of transcription factor protein.
Owner:王进科 +1
Detection of transcription factor protein by double chain RNA molecule with special labeling fixed to micro-plate coated by exonuclease III
InactiveCN1721547ALimit commercializationHigh Throughput Analysis CapabilitiesMicrobiological testing/measurementRegulation of gene expressionTissue extracts
Transcription factor is one important protein for gene regulation, is center of gene expression regulating passage and network, is important target of functional genome and protein block research and important target site of transcription treatment and medicine research. The present invention proposes exonuclease III digesting double stranded nucleic acid molecule coated in microporous plate and containing special marker to detect transcription factor protein. The process of analyzing transcription factor expression and activation level includes the following steps: preparing nucleic acid; fixing the nucleic acid molecules to the pores of the microporous plate; incubation of cell or tissue extract containing transcription factor in the plate; incubation of exonuclease III reaction liquid in the plate; incubation of specific conjugate with special marker on nucleic acid on the plate; detecting and analyzing the specific conjugate to obtain the expression and activation level.
Owner:王进科 +1
Plant bivalent anti-reverse gene bielement expression carrier
InactiveCN1757740AIncrease resistanceAnti agingVector-based foreign material introductionDNA/RNA fragmentationPlant genetic engineeringGenetic engineering
A binary expression carrier of Agrobacterium for culturing the seeds of plant by genetic engineering contains the closely linked corm ubiquitin promoter Pubi regulated Arabidopsisí» cold response gene transcription activation factor gene cbf1 and the Arabidopsis specific senescence gene promote SAG12 regulated iso-pentenyl transferase ipt geneí»s two-valence stress-resistant gene. Its preparing process includes such steps as inserting the T-Nos of sag12-ipt fusion gene into a binary carrier, inserting ubi-cbf1 fusion gene and inserting sag12-ipt fusion gene.
Owner:INST OF FORESTRY CHINESE ACAD OF FORESTRY
Detection method for Apostichopus japonicas AjE101 micro-satellite DNA label
InactiveCN102140522AGood polymorphismImprove stabilityMicrobiological testing/measurementGerm plasmGenetic diversity
The invention belongs to the field of molecular biology DNA labeling technology and application, and in particular relates to Apostichopus japonicas express sequence tag EST micro-satellite label screening development and application. The invention provides Apostichopus japonicas AjE101 micro-satellite specific DNA primers, a polymerase chain reaction (PCR) reaction system by utilizing the primers, and a detection method for an Apostichopus japonicas AjE101 micro-satellite DNA label. The detection method comprises the following steps of: extracting genome of Litopenaeus vannamei Boone; designing the specific primers at two ends of an Apostichopus japonicas AjE101 micro-satellite DNA core sequence; and performing PCR amplification on genome DNA of different groups of the Litopenaeus vannamei Boone or individuals in the group by using the primers, analyzing products, and determining the genome of each individual so as to obtain an Apostichopus japonicas polymorphism genetic variation map. The invention is mainly applied to Apostichopus japonicas germ plasm resource and genetic diversity analysis, cular population genetics, construction of genetic maps and research of functional genes.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Organization chip for researching functional genome as well as preparation method and application thereof
The invention relates to the biochips field, in particular to a tissue chip used for functional genome research and the preparation method as well as the application thereof. The tissue chip of the invention includes a substrate and a paraffin section arranged on the surface of the substrate, wherein, the paraffin section is provided with tissue sample points which are in different development stages and array according to the tissue dynamic development process. Because the tissue chip of the invention can be combined with a gene chip technology, the space-time expression of a difference expressive gene screened out by the chip technology in the tissue dynamic development process is validated and researched in the high-quality, economic and available way.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
Cytochrome P450 enzyme gene for taking part in sanguinarine and chelerythrine synthesis in macleaya cordata and application of cytochrome P450 enzyme gene
ActiveCN106119265AAchieve synthesisAchieve in vitro synthesisTransferasesOxidoreductasesSanguinarineChelerythrine
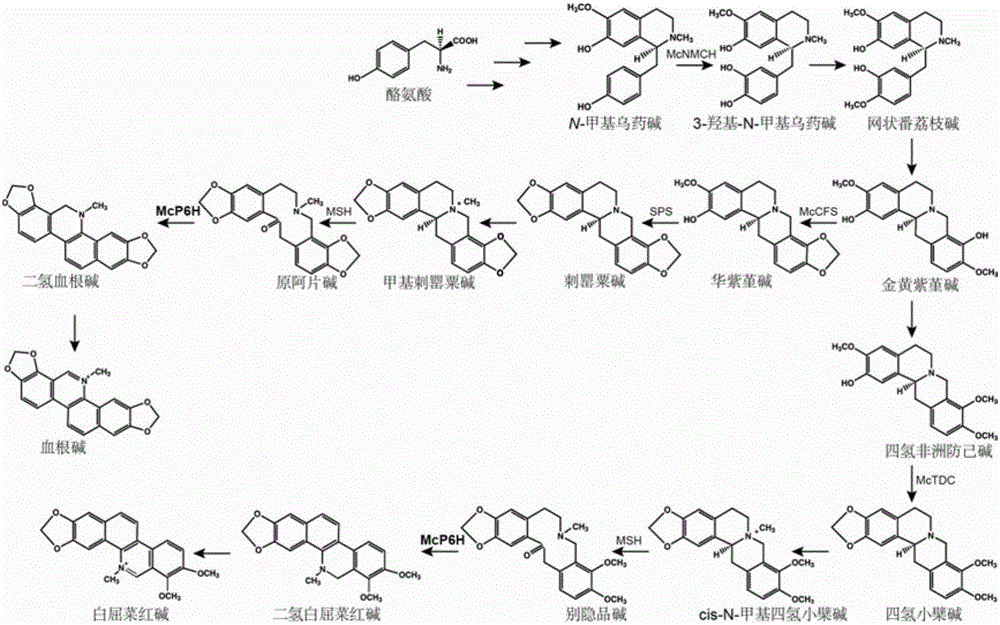
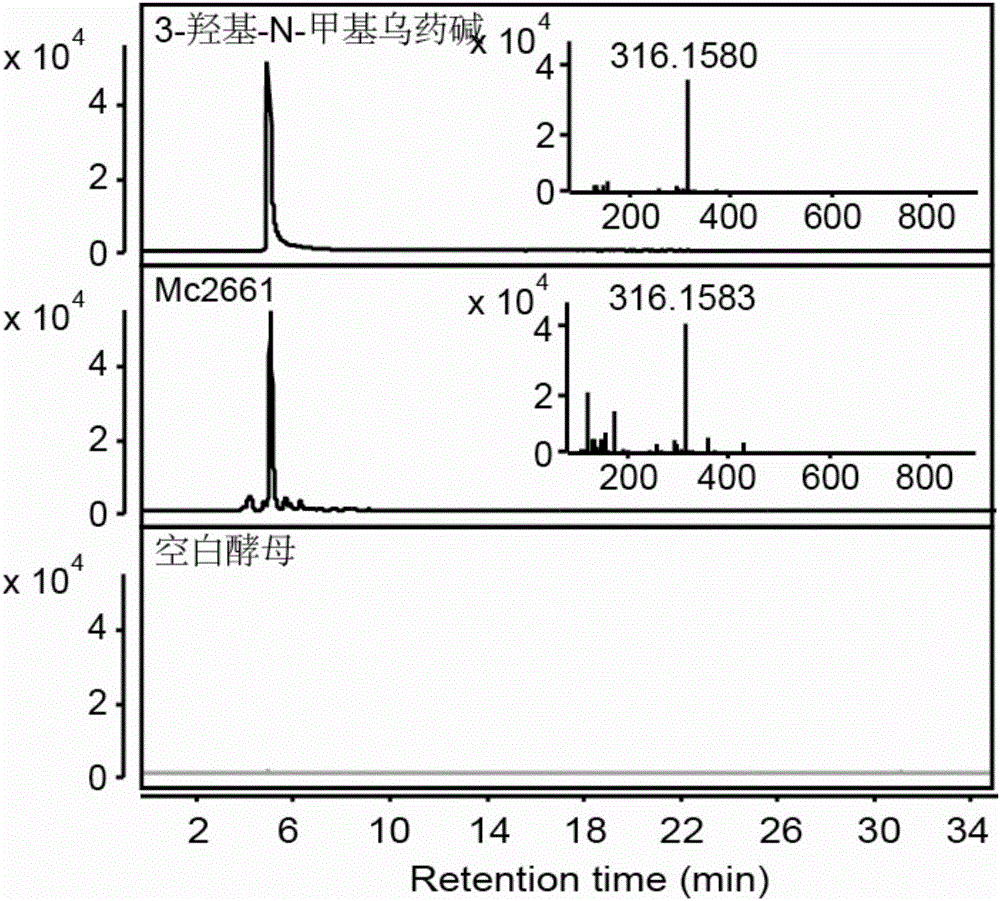
The invention discloses cytochrome P450 enzyme gene for taking part in sanguinarine and chelerythrine synthesis in macleaya cordata and application of the cytochrome P450 enzyme gene. For the first time, five cytochrome P450 enzyme genes taking part in sanguinarine and chelerythrine synthesis are found in macleaya cordata genome and comprise gene Mc9485, gene Mc2661, gene Mc217, gene MC11229 and gene Mc11218. A brewer's yeast system verifies that the steps of reaction are respectively verified by upstream precursor feeding and synthesis of sanguinarine and chelerythrine intermediate is achieved. According to the cytochrome P450 enzyme gene, the conversion efficiencies of the same function genes in the macleaya cordata, the papaver somniferum and the eschscholzia californica are compared by the brewer's yeast heterogeneous expression system, and the conversion efficiency of the macleaya cordata is obviously higher than those of the papaver somniferum and the eschscholzia californica. According to the cytochrome P450 enzyme gene, a molecular mechanism of the sanguinarine synthesis in the macleaya cordata is further revealed. Thus, a theoretical basis and a molecular assistant breeding target are provided for the breeding of the macleaya cordata with high sanguinarine and chelerythrine content. Meanwhile, valuable experience is provided for in-vitro artificial synthesis of the sanguinarine and the chelerythrine.
Owner:MICOLTA BIORESOURCE INC LTD
Major QTL and SNP molecular marker for controlling Nelumbo nucifera color traits as well as detection primer and application thereof
ActiveCN111979349AHigh contribution rateSpeed up the breeding processMicrobiological testing/measurementAgainst vector-borne diseasesElectrophoresesGenotype
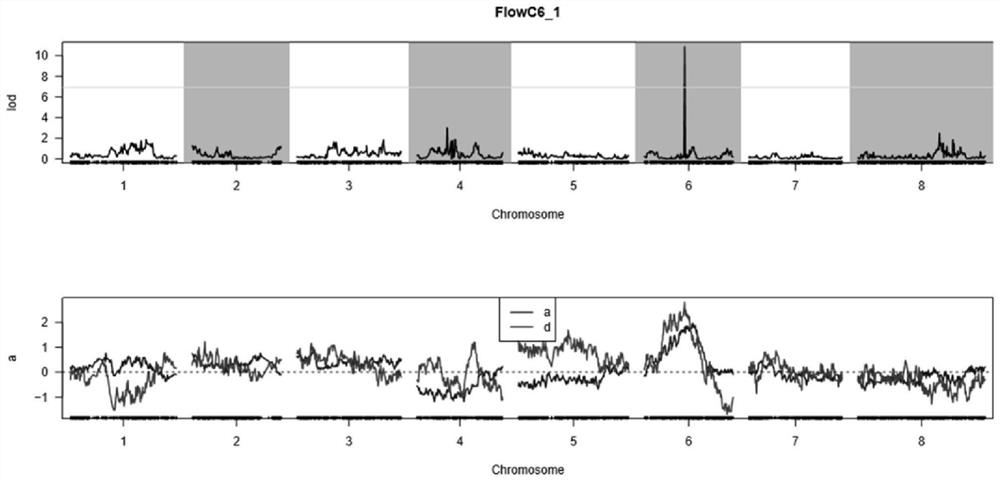
The invention discloses a major QTL and SNP molecular marker for controlling Nelumbo nucifera color traits as well as a detection primer and application thereof. The invention firstly provides a majorQTL site for controlling the flower color traits of Nelumbo nucifera, and the major QTL site is positioned in a sixth linkage group and between two SNP markers; the major QTL is closely linked with the SNP molecular marker, has a relatively high contribution rate to the flower color traits of the Nelumbo nucifera, participates in regulating and controlling the phenotype of the red or white flowerof the Nelumbo nucifera, and can be used for map-based cloning mining of functional genes for controlling the flower color traits and molecular marker-assisted selection. The invention further provides an SNP molecular marker closely linked with the major QTL, and the SNP molecular marker is used for efficiently selecting Nelumbo nucifera varieties with target flower colors and molecular marker assisted breeding of Nelumbo nucifera with seeds and Nelumbo nucifera with roots. The invention further provides a PARMS primer group for detecting the SNP marker and a method for identifying the red and white flower traits of the Nelumbo nucifera, so that the flower color genotype data of the Nelumbo nucifera can be analyzed and obtained only through PCR amplification and without electrophoresis detection.
Owner:WUHAN ACADEMY OF AGRI SCI
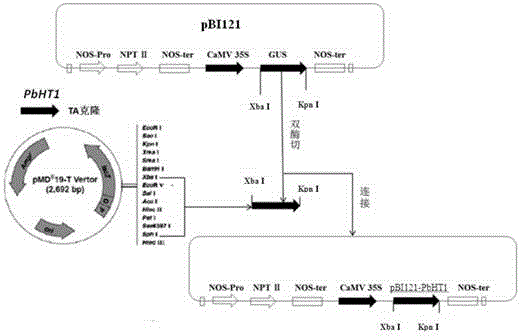
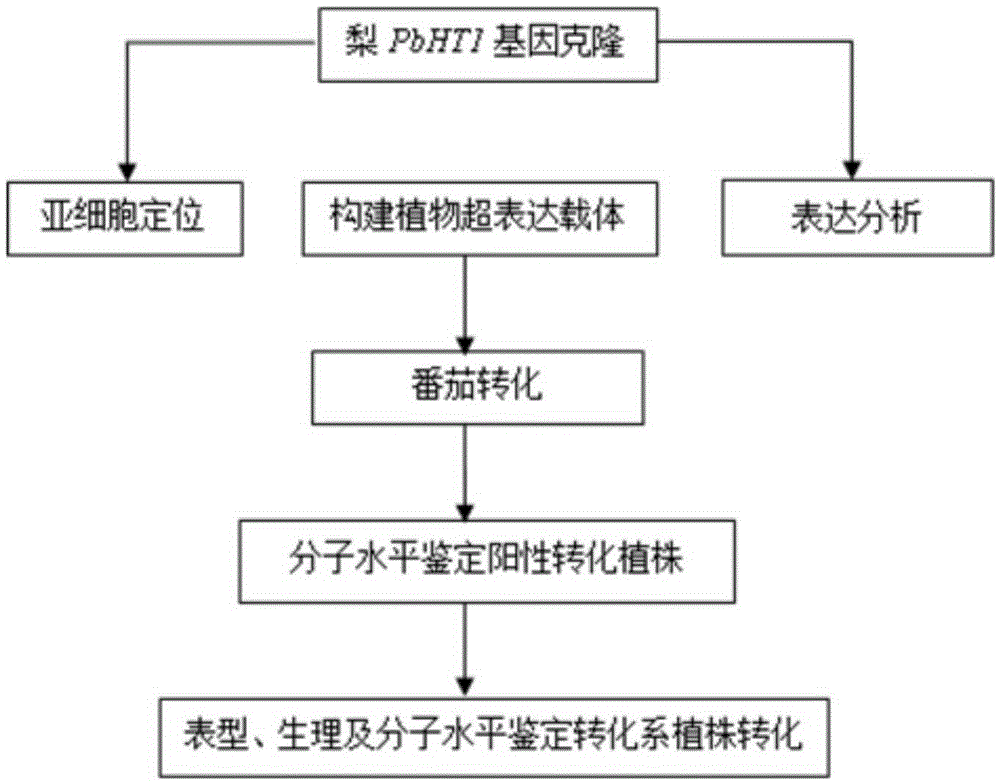
Pear hexose transport protein gene PbHT1 and application thereof
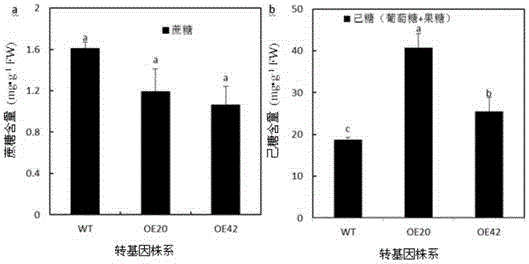
ActiveCN105296502AImprove floweringIncrease fruit hexose contentPlant peptidesFermentationBiotechnologyPEAR
The invention discloses a pear hexose transport protein gene PbHT1 and application thereof. The hexose transport protein gene PbHT1 is separated from a pear fruit, the nucleotide sequence of the gene is as indicated in SEQ ID No. 1, and the coded amino acid sequence of the gene is as indicated in SEQ ID No. 2. The gene PbHT1 is transformed into a tomato through a southern buddhism genetic transformation method, a transgenic plant is obtained, and through biological function verification, it is shown that the clonal gene PbHT1 has the effects of postponing the flowering phase and raising the hexose content in the fruit. Through finding of the gene PbHT1, a new gene resource is provided for molecular breeding for improving quality of the plant fruit.
Owner:NANJING AGRICULTURAL UNIVERSITY
Method for judging strain participating in antimony reduction process in soil and key function gene of strain
ActiveCN110317863ASimple structureBacteriaMicrobiological testing/measurementBiotechnologyMicroorganism
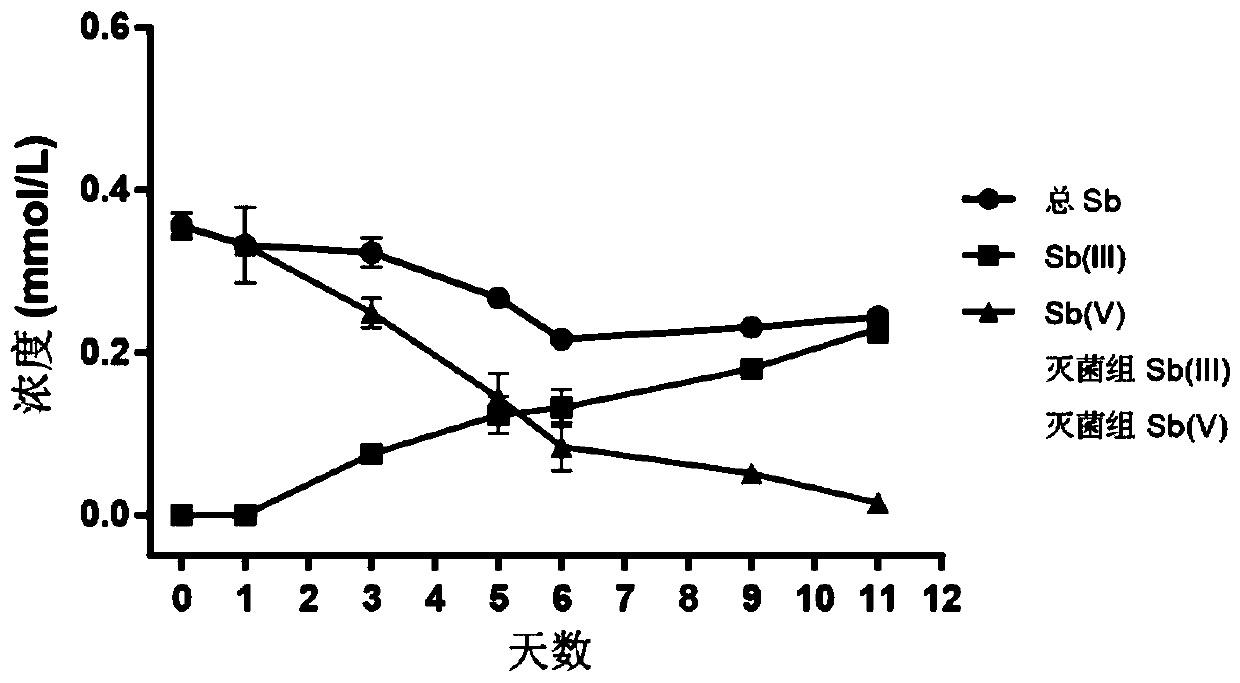
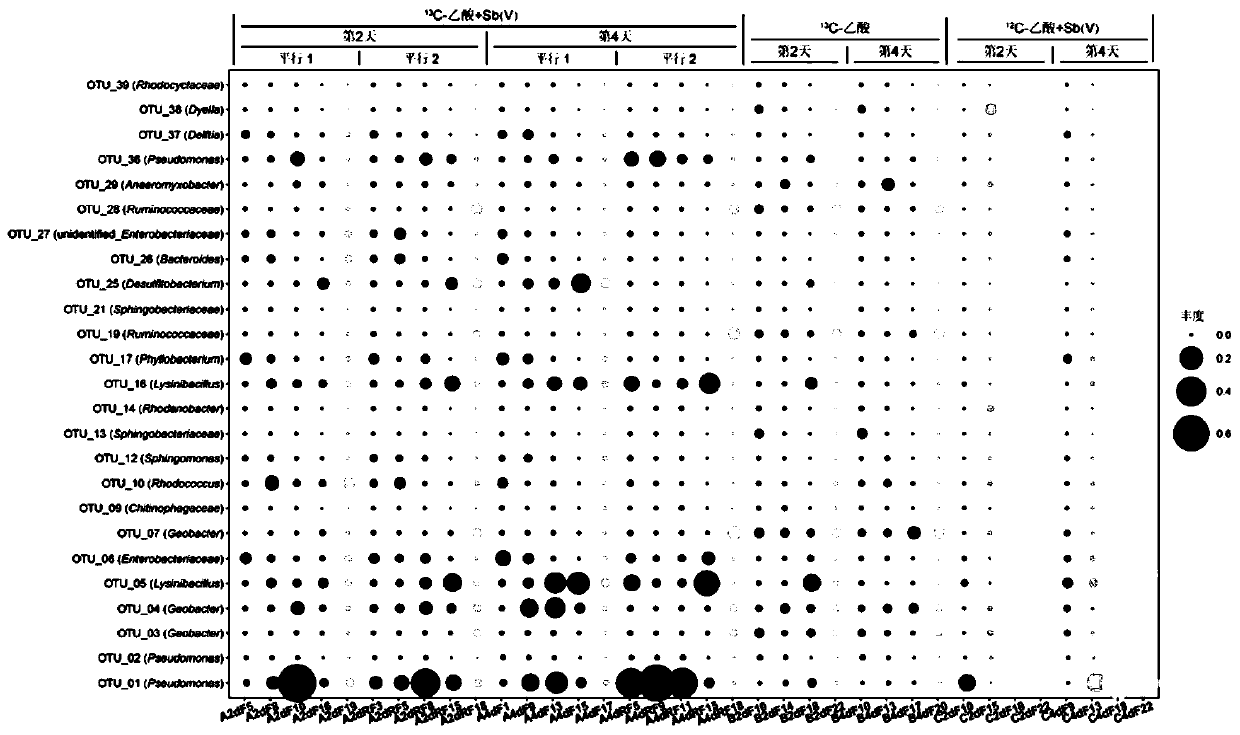
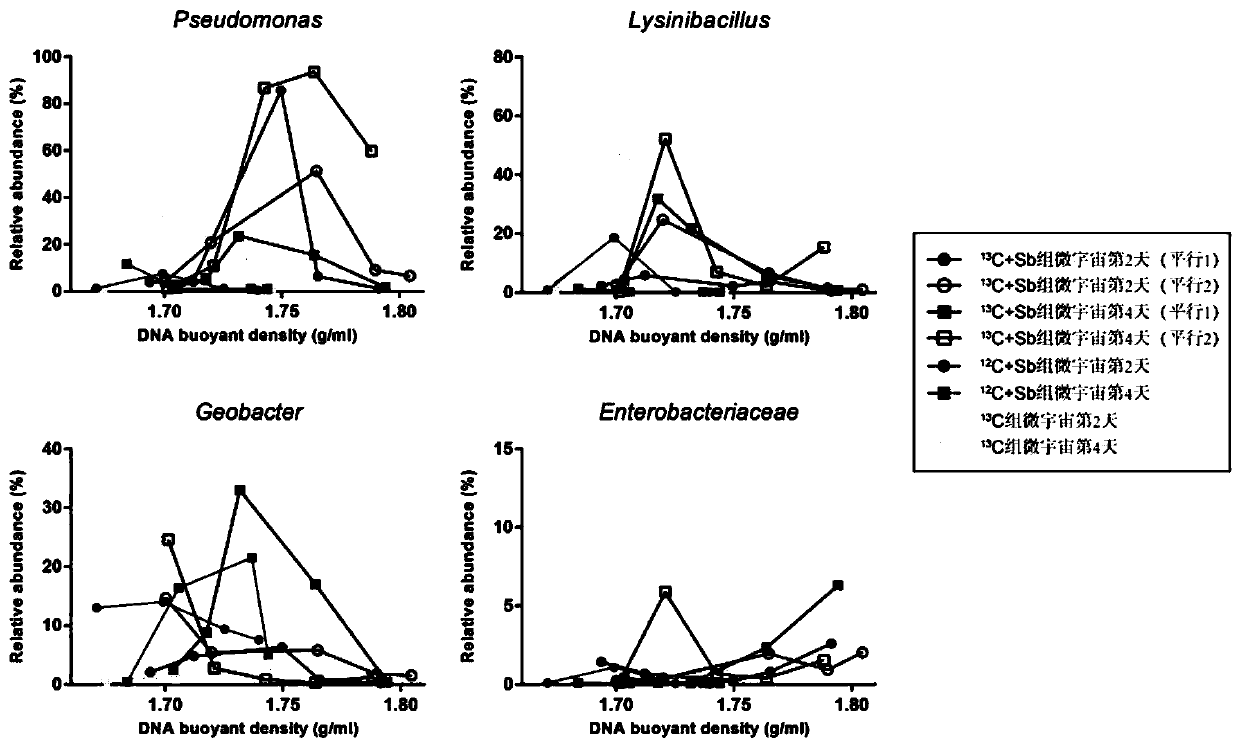
The invention discloses a method for judging a strain participating in the antimony reduction process in soil and a key function gene of the strain. After a substrate of the strain is consumed throughstarved culture, a unique metabolism substrate is added, a unique electron acceptor Sb(V) is provided, so that only one leading electron exchange process exists in a system, a microorganism metabolizes and oxidizes the organic substrate, antimony reduction is driven through coupling, and accordingly the Sb(V) obtains electrons and is reduced into an Sb(III). According to the method, the Sb(V) reduction phenomenon of an anaerobic culture system, subjected to Sb(V) stress, of the rice field soil is observed, and a DNA-SIP technology is used for identifying phylogeny information of the microorganism which can drive the Sb(V) reduction process in the culture system. According to the method, metabolism information of the antimony reduction function microorganism class group and the key function microorganism in the rice field soil is mined, and the method has important significance on knowing the antimony reduction process driven by the microorganism and cognizing antimony reduction bacteria and the key function gene.
Owner:GUANGDONG INST OF ECO ENVIRONMENT & SOIL SCI
Method for developing polymorphic SSR markers based on transcriptome sequences
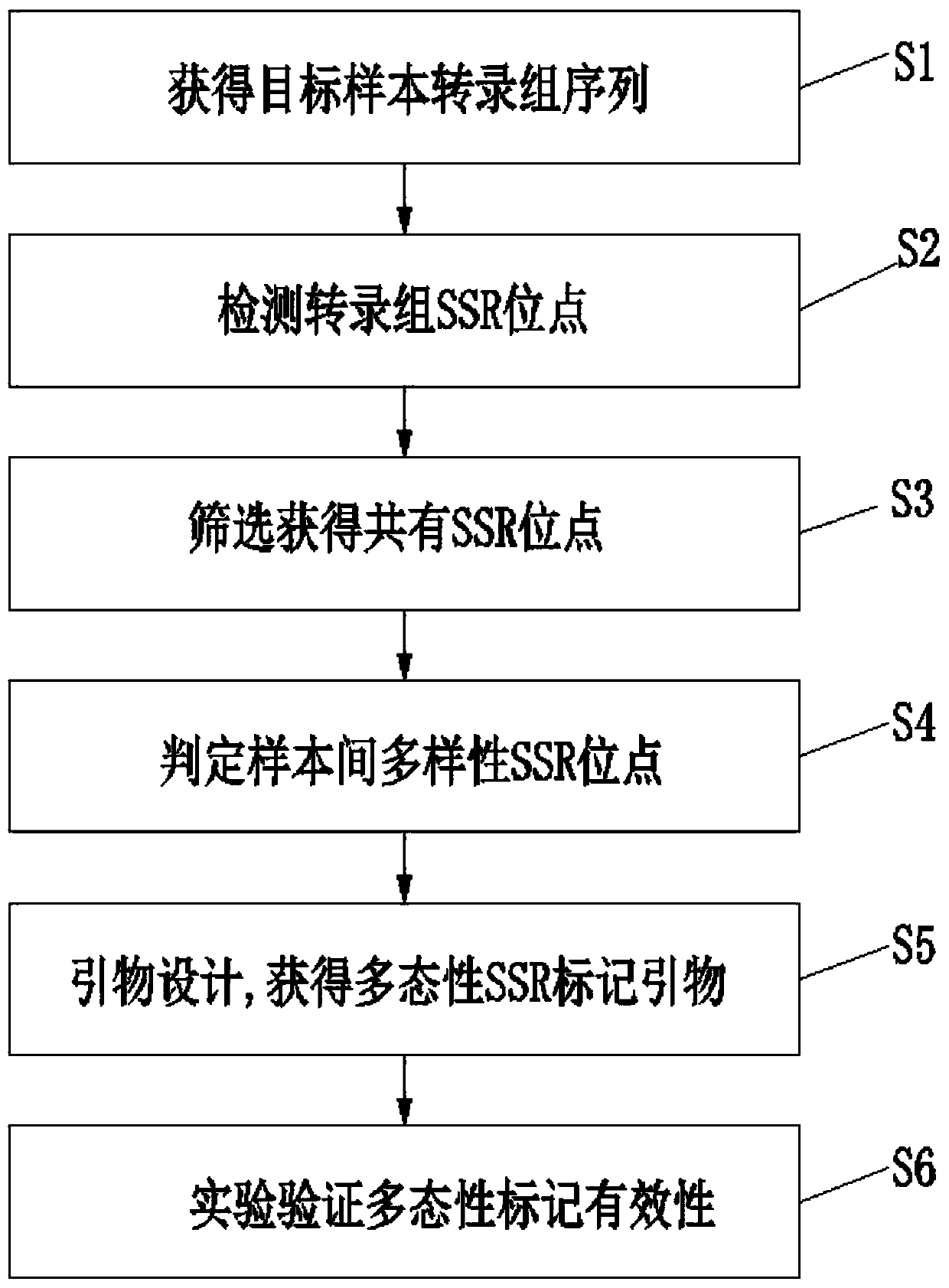
InactiveCN110669834AVerify validityImprove development efficiencyMicrobiological testing/measurementProteomicsContigConserved sequence
The invention discloses a method for developing polymorphic SSR markers based on transcriptome sequences. The method comprises the following steps: S1, the transcriptome sequences of a plurality of samples of a target species are obtained; S2, potential SSR site information is detected; S3, the types of SSR locus repeats and adjacent nucleotide sequence information of the SSR locus repeats are obtained by screening; S4, polymorphic SSR candidate loci among the samples are screened; and S5, transcriptome sequence sets carrying the polymorphic SSR loci are spliced into a non-repetitive contig Uni-contig, primers are designed according to a side wing conservative sequence of the non-repetitive contig Uni-contig, and marker polymorphic detection is conducted. The polymorphic SSR markers can beeffectively screened through the transcriptome sequences, the configuration requirement for a computer is reduced, the method can be completed by a common computer, the effectiveness of the molecularmarker primers is effectively improved, the development time is shortened, the development cost is reduced, meanwhile, the PCR amplification fragment length of the polymorphic SSR markers and linkagegene function information of the polymorphic SSR markers can further be predicted, the development efficiency of the transcriptome SSR markers is remarkably improved, and molecular assisted breedingand functional gene research are promoted.
Owner:INST OF FOOD CROPS HUBEI ACAD OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com