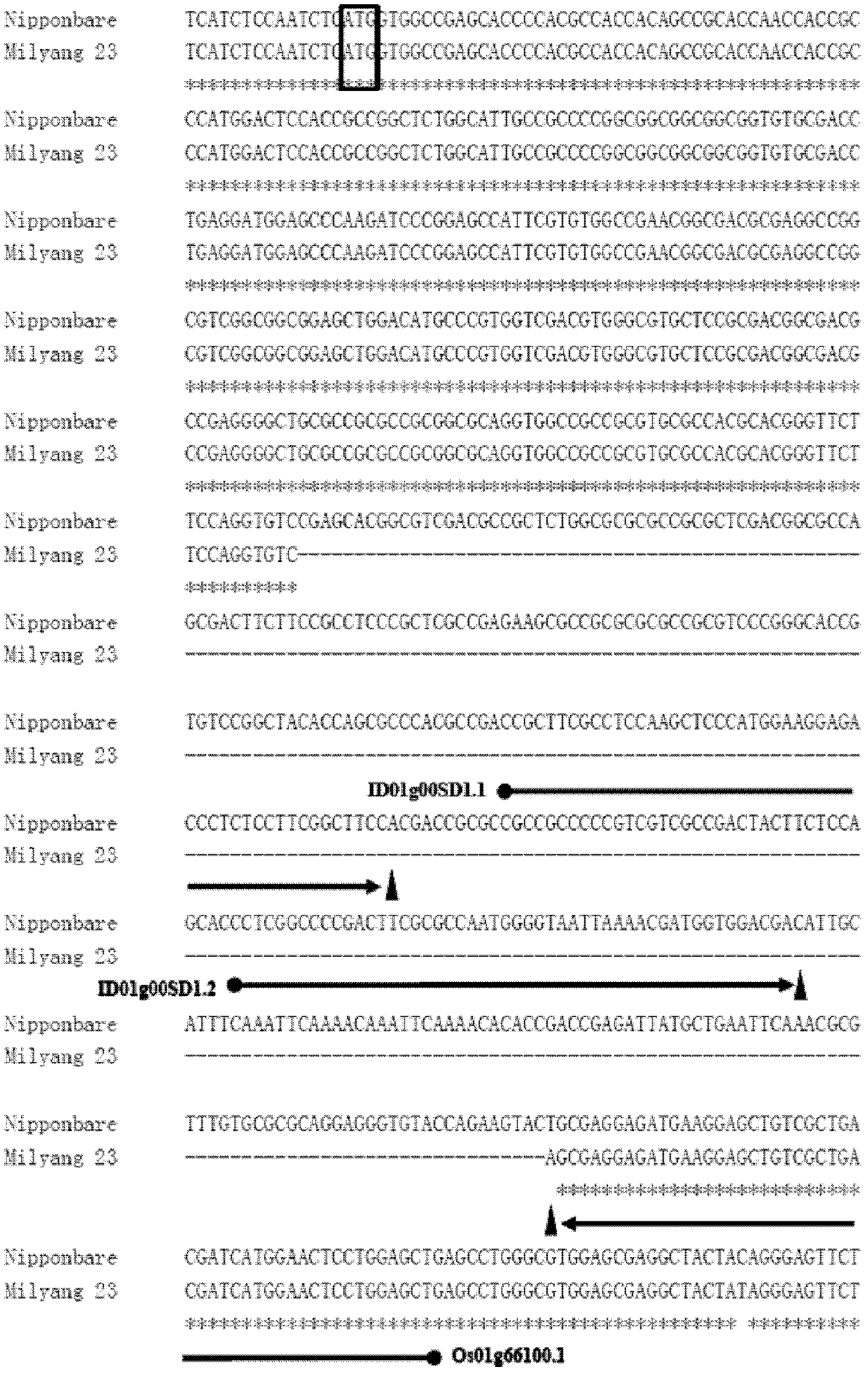
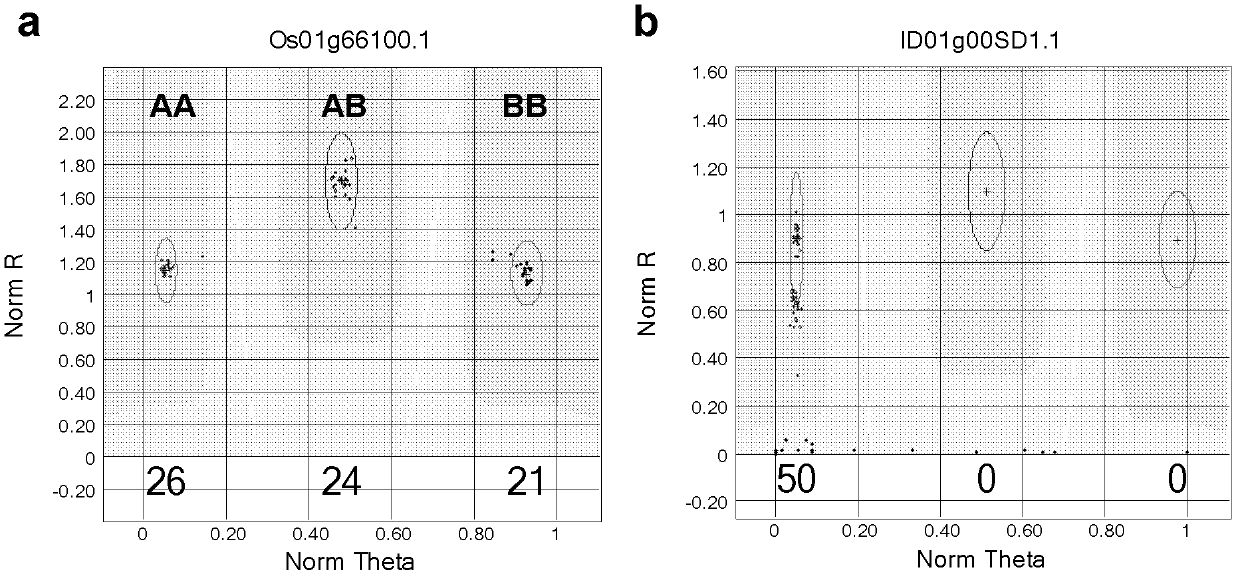
Rice whole genome SNP chip and application thereof
A whole genome and chip technology, applied in the field of rice whole genome SNP chip and preparation, rice SNP chip field, can solve the problems of limited use, high data analysis requirements, no chip, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Embodiment 1: RICE6K SNP chip preparation method
[0055] 1. Obtaining the first type of probes on the RICE6K SNP chip
[0056] The applicant used the Illumina next-generation sequencing technology to sequence the genome of the rice core parent material. In addition, at the end of 2010, Huang et al. published genome resequencing data of 520 rice landraces (Huang et al., Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010, 42: 961-967). The applicant downloaded this data from a public database and presented it as the Nipponbare Genome (MSU version 6.1, http: / / rice.plantbiology.msu.edu / ) as the reference sequence, using MAQ software ( http: / / sourceforge.net / projects / maq ) to match all species resequencing data to the reference sequence. The specific steps of probe design are as follows:
[0057] (2) First, 4,236,029 high-quality SNPs were identified according to the following criteria:
[0058] Sequences were used to identify ...
Embodiment 2
[0075] Example 2: Application of RICE6K Rice SNP Chip in Detection of Rice DNA Samples
[0076] 1. Rice genomic DNA extraction: Genomic DNA is extracted from rice seeds, leaves and other tissues according to the detection requirements. Among them, the DNA of young rice leaves was extracted using the Promega Plant Genome Extraction Kit according to the standard procedure (Wizard Magnetic 96 DNA Plant System Kit, Cat. No. FF3760 or FF3761, Promega, USA).
[0077] 2. DNA sample quality detection: detect with agarose gel electrophoresis with a mass fraction of 1% (W / W), use a gel imaging system (Gel Doc XR System, Bio-Rad, USA) to judge the electrophoresis results, and ensure that the genomic DNA The integrity is good, and the length of the genomic DNA fragment is greater than 10kb; use a UV spectrophotometer (NanoDrop2000, Thermo Scientific, USA) to measure the protein and organic matter pollution levels in the genomic DNA, and the genomic DNA A260 / 280 ratio should be between 1.8...
Embodiment 3
[0080] Example 3: Application of RICE6K Rice SNP Chip in Molecular Marker Fingerprint Analysis of Rice Germplasm Resources
[0081] Using the RICE6K SNP chip, the applicant conducted molecular marker fingerprint analysis on 106 micro-core rice germplasm resources. The RICE6K chip detected the genomic DNA of these 106 rice varieties. Among the 5,102 effective detection sites, 636 sites were detected as heterozygous sites in more than 3 varieties. The applicant believes that the detection ability of these sites is relatively low. Poor, the remaining 4466 high-quality loci were used for genotyping of 106 varieties. According to the genotyping results, the cluster analysis of these 106 rice varieties showed that all 18 japonica rice varieties were clustered into one category, and the other 88 indica rice varieties were clustered into another category. The applicant analyzed the number of polymorphic SNP sites between any two varieties that could be detected by this batch of RICE6...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com