Method for developing polymorphic SSR markers based on transcriptome sequences
A transcriptome sequence and polymorphism technology, applied in the field of molecular biology, can solve problems such as high dependence on computer configuration, poor operability, and large amount of calculation, so as to shorten development time and cost, improve effectiveness, and reduce configuration required effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0026] The technical solutions of the present invention will be clearly and completely described below in conjunction with the accompanying drawings of the present invention. Based on the embodiments of the present invention, all other embodiments obtained by persons of ordinary skill in the art without creative efforts fall within the protection scope of the present invention.
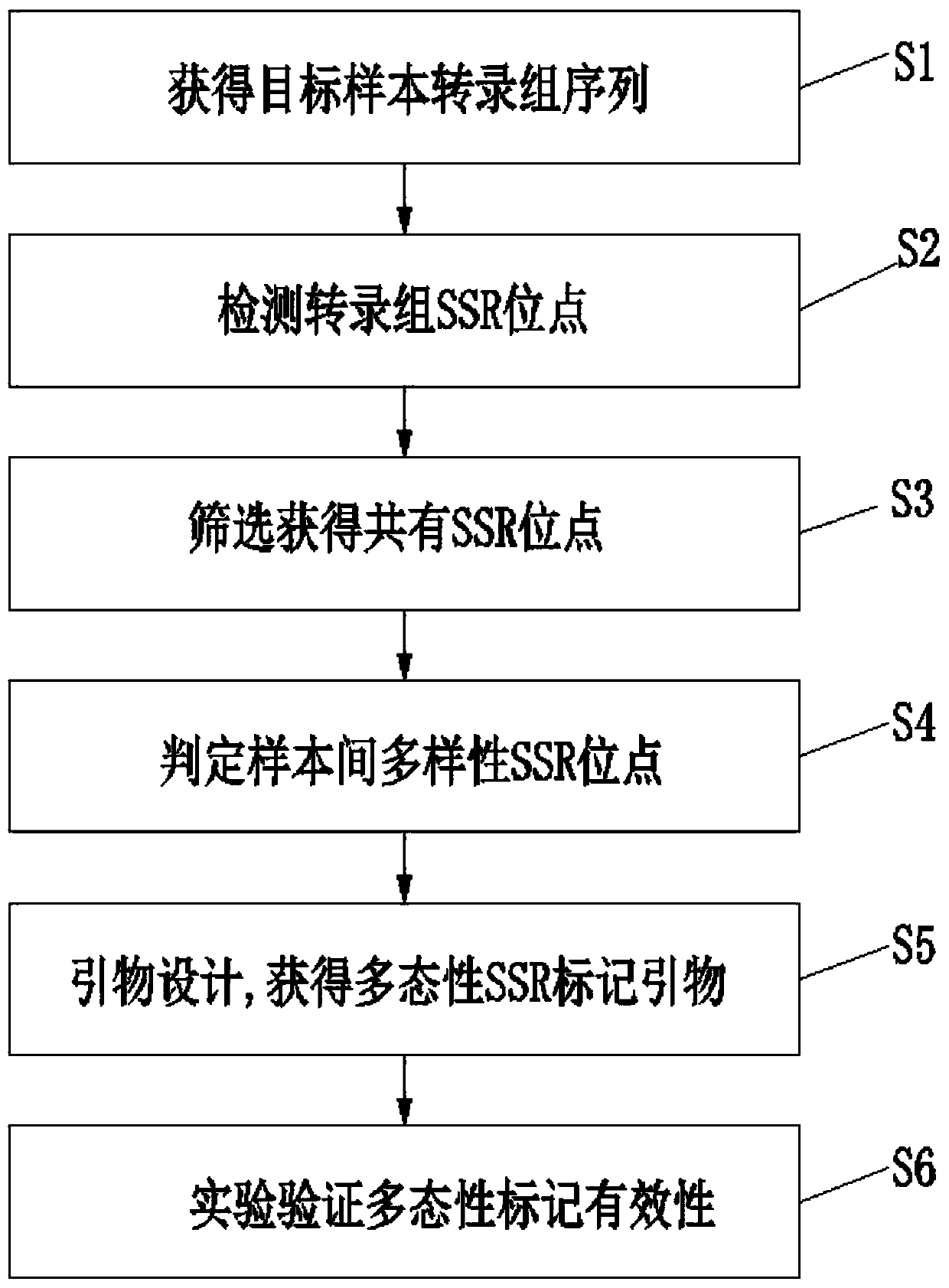
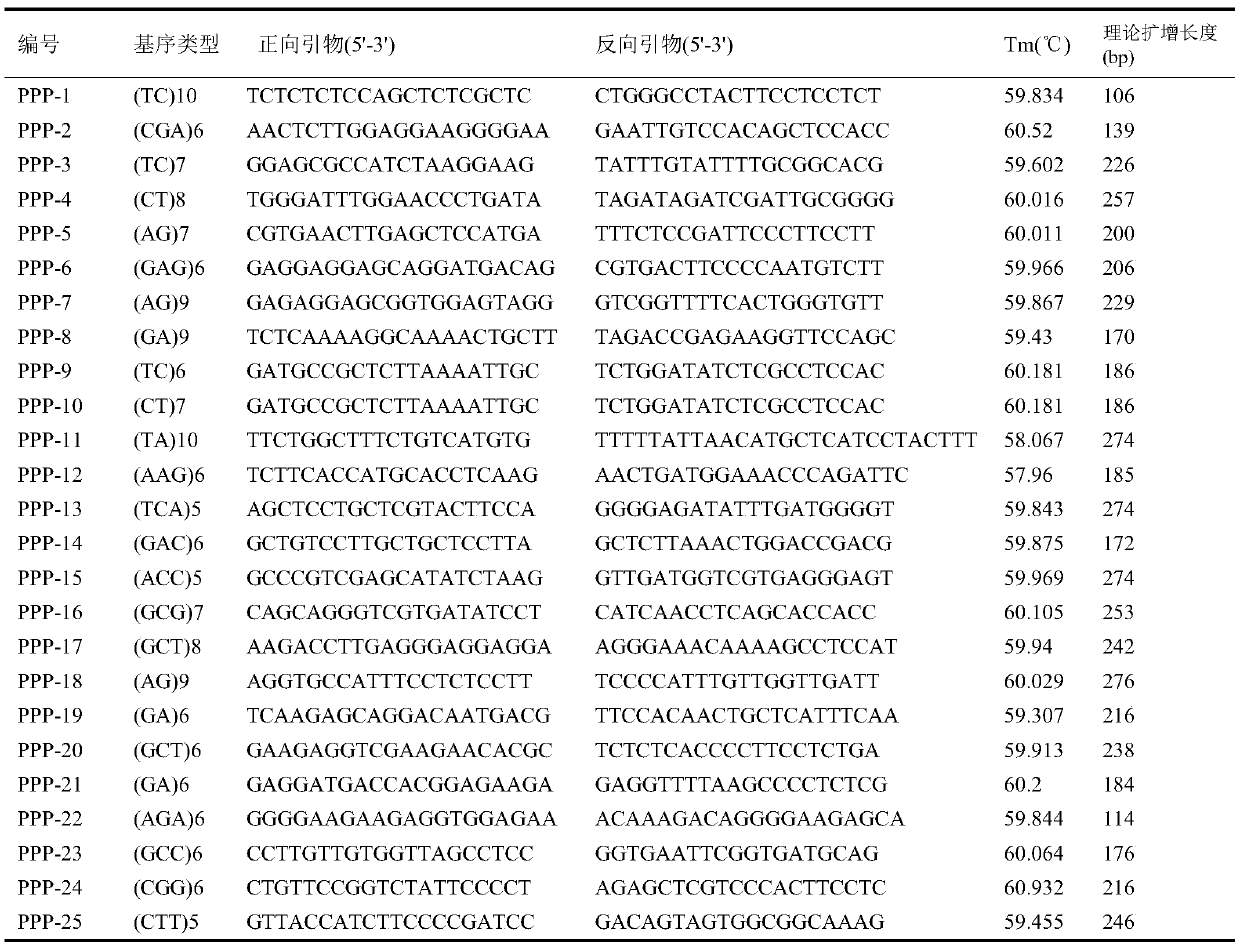
[0027] Such as figure 1 , figure 2 , the application provides a method for developing polymorphic SSR markers based on transcriptome sequences, comprising the following steps:
[0028] S1. First, take several samples of the target species, perform high-throughput sequencing on their transcriptomes to obtain Reads, and remove low-quality sequencing Reads (the number of bases with a quality value Q≤5 accounts for more than 50% of the entire read), so as to ensure Obtain high-quality transcriptome sequences, and then use Trinity software to assemble all the reads obtained by sequencing, and finally ob...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com