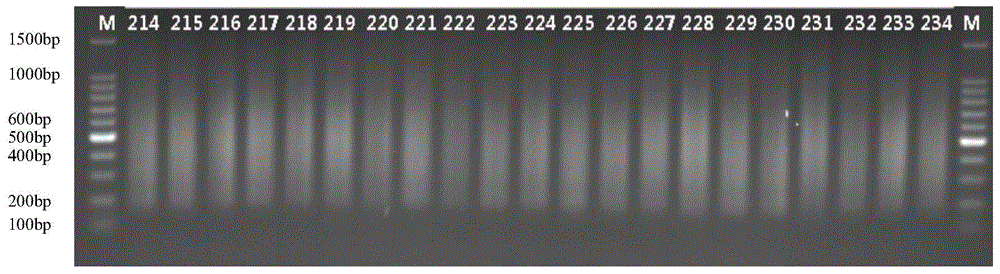
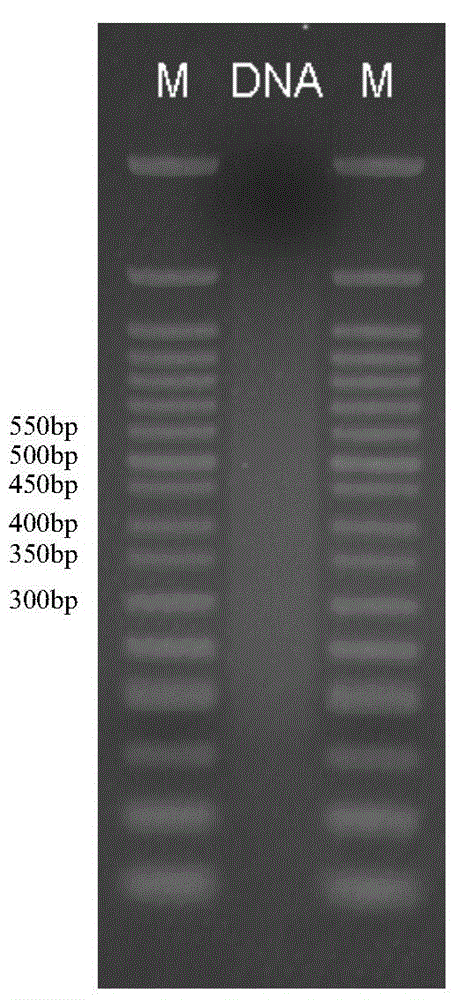
Method for constructing high-flux simplified genome sequencing library
A genome sequencing and construction method technology, applied in the field of high-throughput simplified genome sequencing library construction, can solve the problems of large amount of genomic DNA, discarding original data, and high cost of library construction, achieving high throughput, reducing impact, and simplifying construction The effect of the process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 High-throughput Simplified Genome Sequencing Library Construction of the Genetic Map of Mei F1 Population
[0044] 387 offspring of the Prunus mune F1 population and their two parents were selected as experimental materials, and the construction of a high-throughput simplified genome sequencing library included the following steps:
[0045] Step 1: Genomic DNA of 387 progenies of Mei F1 population and their two parents was diluted to 50ng / μL for later use; genomic DNA fragmentation: the diluted genomic DNA was digested with blunt-end restriction endonucleases HaeIII and Hpy166II, The enzyme digestion reaction system is:
[0046]
[0047] The mixed system was incubated in a constant temperature water bath at 37°C for 5 hours.
[0048] Step 2: Add the reaction reagents required to generate the 3' cohesive A-terminus to the product of the 1st-step enzyme digestion reaction, including:
[0049]
[0050] The mixed system was incubated in a constant temperatur...
Embodiment 2
[0090] Example 2 Construction of High-throughput Simplified Genome Sequencing Library of Flax Genetic Map
[0091] In this embodiment, 115 offspring of the flax F2 population and their two parents were selected as experimental materials, and the genomic DNA was diluted according to the first step in Example 1; the genomic DNA fragment was double-digested with RsaI and HaeIII, and the reaction system was the same as in Example 1; Add A to the 3' end according to step 2 in Example 1; connect each sample with a barcode adapter according to step 3 in Example 1; perform PCR reaction according to step 4 in Example 1 (primers for PCR reaction and conditions are the same as in Example 1); the PCR product is purified and detected; the PCR purified products of 2 parents and 115 progeny are mixed according to a mass ratio of 40:8 and cut into gel to select fragments, and a fragment of 450-550bp is selected; according to the embodiment In step 5 of 1, perform low-cycle PCR amplification a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com