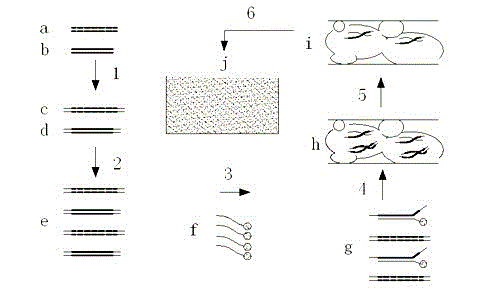
Novel capture and enrichment technology for targeting nucleic acid molecules
A technology targeting nucleic acids and molecules, which is applied in the field of capture and enrichment of nucleic acid molecules, can solve the problems of high probe labeling price, high capture cost, connection efficiency affecting capture and enrichment yield, etc., to improve detection sensitivity, Improve hybridization efficiency and increase the effect of collision chance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 Using polyacrylamide gel capture technology and high-throughput sequencer to analyze the distribution of various functional genes in the intestinal microbial metagenome:
[0045] (1) Design and preparation of probes
[0046] Selection of DNA sequence fragments of functional genes of intestinal microorganisms: use software such as Mega or Clustal W to perform bioinformatics comparison and complete the selection of 20,000 functional gene nucleic acid sequences.
[0047] Probe design: use software such as primer 5.0 to design probes for specific fragments of microorganisms.
[0048] Synthesis and labeling of probes: Probes were synthesized using a DNA synthesizer, and after all probes were synthesized, an acrylamide group was labeled at the 5' end.
[0049] (2) Intestinal microbial community metagenomic extraction, genome fragmentation and adapters on both sides
[0050] Intestinal microbial community metagenomic DNA extraction: collect 200mg of fresh stool, ...
Embodiment 2
[0064] Example 2 Targeted analysis of exon mutations in cancer-related genes in the human genome using polyacrylamide gel capture and high-throughput sequencing:
[0065] (1) Design and preparation of probes
[0066] Exon nucleic acid sequence selection of cancer-related genes: use software such as Mega or Clustal W to perform bioinformatics comparison and complete the selection of 1,000 functional gene nucleic acid sequences.
[0067] Design of probes: use primer5.0 and other software to design probes for specific fragments of microorganisms.
[0068] Synthesis and labeling of probes: Probes were synthesized using a DNA synthesizer, and after all probes were synthesized, an acrylamide group was labeled at the 5' end.
[0069] (2) Intestinal microbial community metagenomic extraction, genome fragmentation and adapters on both sides
[0070] Human genomic DNA extraction: Collect 2ml of venous blood, lyse red blood cells and digest with proteinase K, and extract human genomi...
Embodiment 3
[0084] Example 3 Targeted analysis of trace pathogenic microorganisms in food using polyacrylamide gel capture and high-throughput sequencer:
[0085] (1) Design and preparation of probes
[0086] Species-specific DNA sequence fragment selection: use software such as Mega or Clustal W to perform bioinformatics comparison and complete the selection of more than 100 specific genes of food pathogenic microorganisms.
[0087] Probe design: use software such as primer 5.0 to design probes for specific fragments of microorganisms.
[0088] Synthesis and labeling of probes: The probes were synthesized using a DNA synthesizer, and an acrylamide group was labeled at the 5' end.
[0089] (2) Genome extraction of pathogenic microorganisms, genome fragmentation and joints on both sides
[0090] Genomic DNA extraction of pathogenic microorganisms: collect chilled meat and duck meat products after slaughtering and processing, and use sterile cotton swabs to conduct six-sided 50cm 2 Af...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com