Efficient specific sgRNA recognition site guide sequence for pig gene editing and screening method thereof
A technology for identifying sites and guide sequences, applied in the fields of genomics and bioinformatics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1 Screening method for efficient and specific sgRNA recognition site guide sequences for swine gene editing
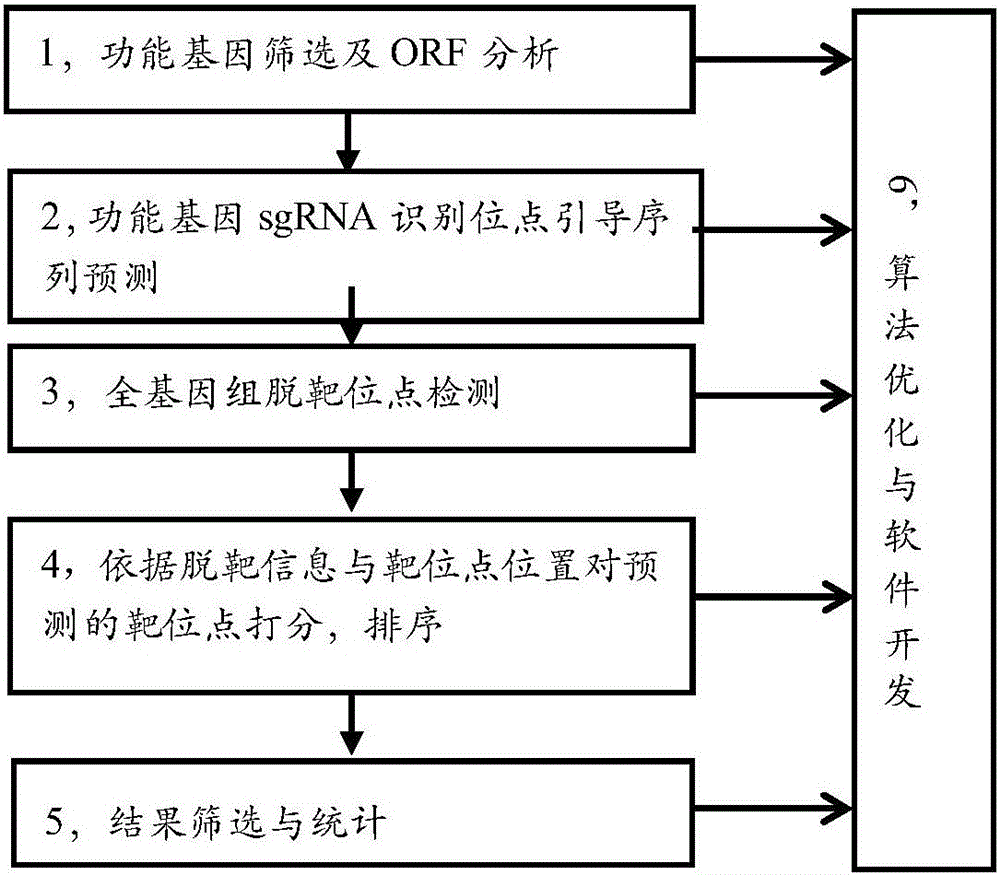
[0029] see figure 1 , the flow chart of the screening method for the efficient and specific sgRNA recognition site guide sequence for pig gene editing in this embodiment, the experimental sample in this embodiment is the genome of pig (Sus scrofa Duroc) that has been sequenced (version 10.2) ) splicing length is 2.8Gb. Since this example is a sequenced species, the SNP correction process is omitted; if the experimental samples are sequenced Duroc pigs, Wuzhishan pigs and Tibet wild boars of sequenced species, the reference genome of the sequenced species can be directly used;
[0030] The screening method for the efficient and specific sgRNA recognition site guide sequence for swine gene editing includes the following specific steps:
[0031] 1) Classification and screening of protein-coding genes in the pig genome
[0032] In the Ensembl (www.ensembl.o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com