High-flux single-cell whole genome amplification method
A whole-genome amplification, single-cell technology, applied in the field of high-throughput single-cell whole-genome amplification, can solve the problems of poor fidelity, non-specific amplification, poor repeatability, etc., and achieve the effect of automation and scale
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
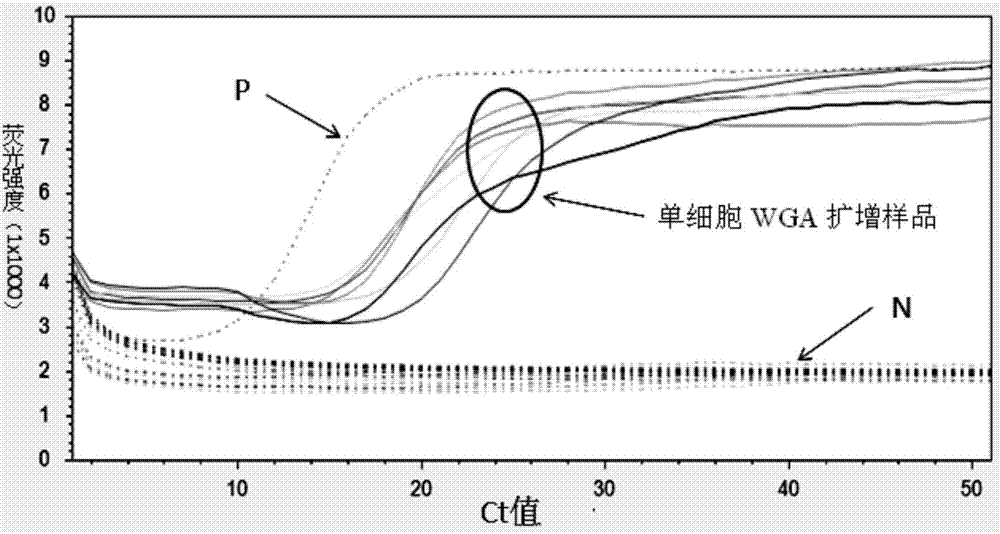
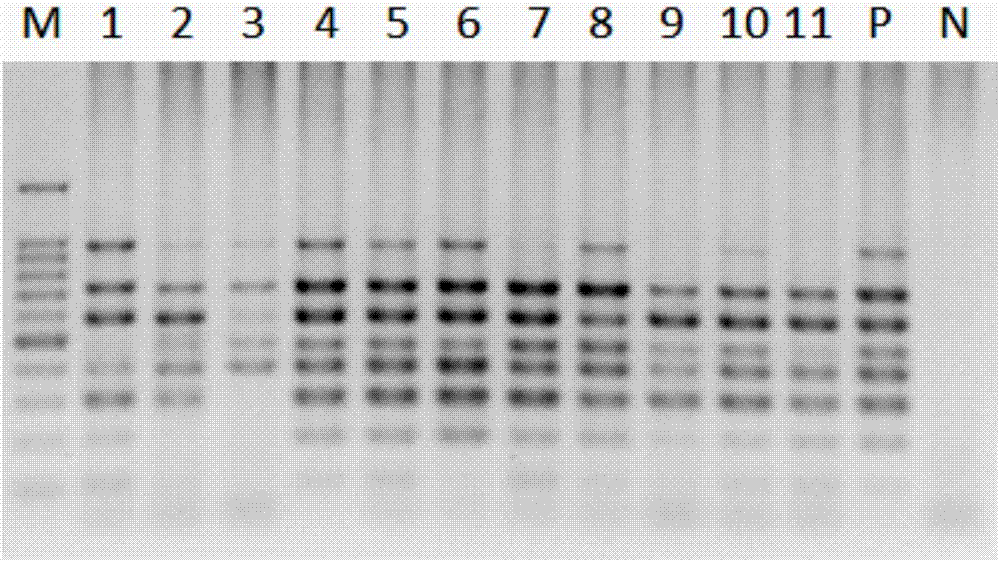
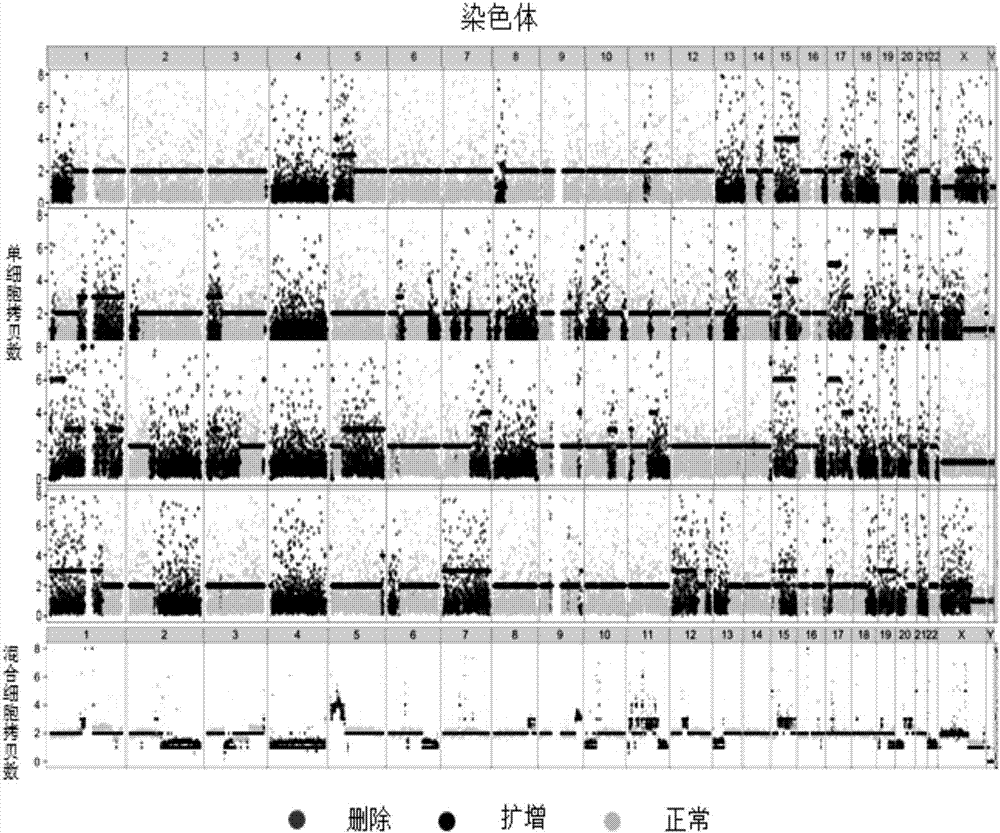
[0021] The present invention will be further described in detail below through specific embodiments in conjunction with the accompanying drawings.
[0022] The high-throughput single-cell whole-genome amplification method of the present invention is realized based on the micro-dispensing platform, because to realize the distribution of single cells, the minimization of samples is very critical. At present, the dispensing ability of the micro-dispensing platform has achieved a nano-upgrade breakthrough, which is a qualitative leap compared with the traditional micro-upgraded micro-dispensing platform. The present invention realizes the distribution of single cells on the basis of the nano-liter micro-dispensing platform, and then realizes a series of in situ cell lysis, neutralization reaction and whole genome amplification reaction, etc., and completes the whole genome amplification of single cells.
[0023] Compared with the prior art, the significant advantages of the presen...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com