Method for identifying type and site of RNA binding with protein in plant
A plant and protein technology, applied in biochemical equipment and methods, microbial assay/inspection, etc., can solve problems such as limiting the function of RNA-binding proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
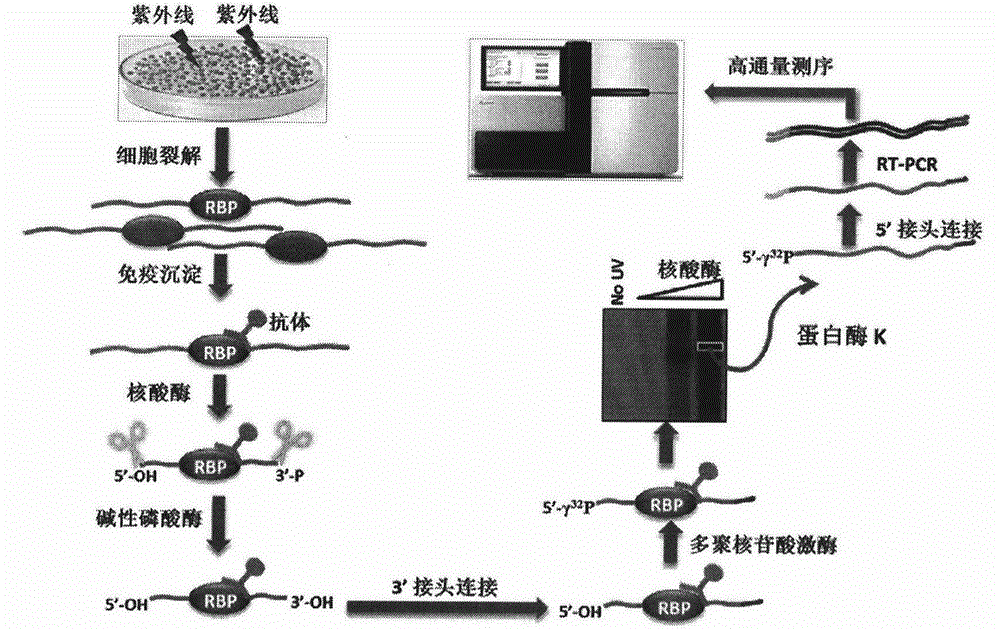
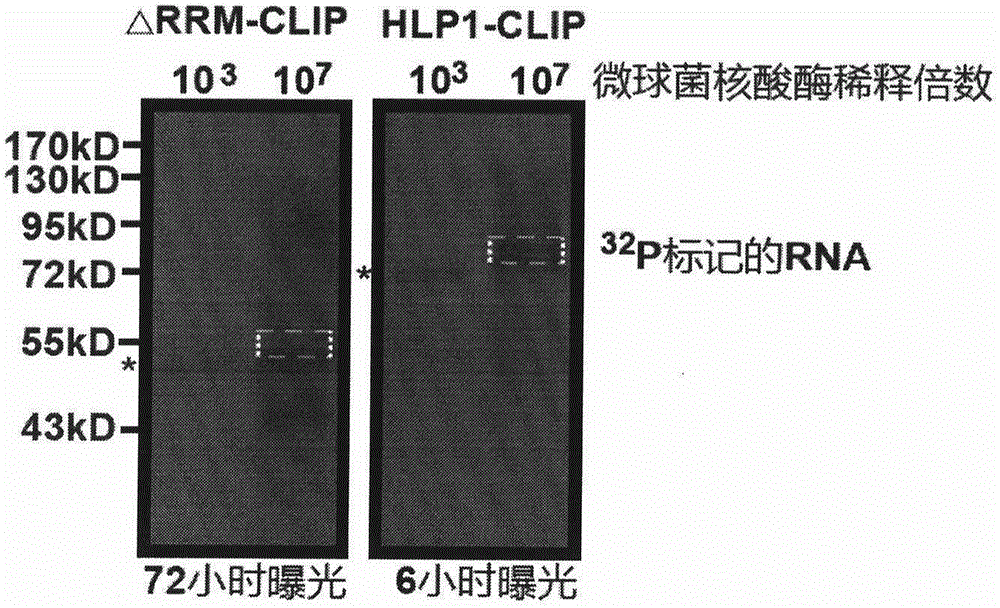
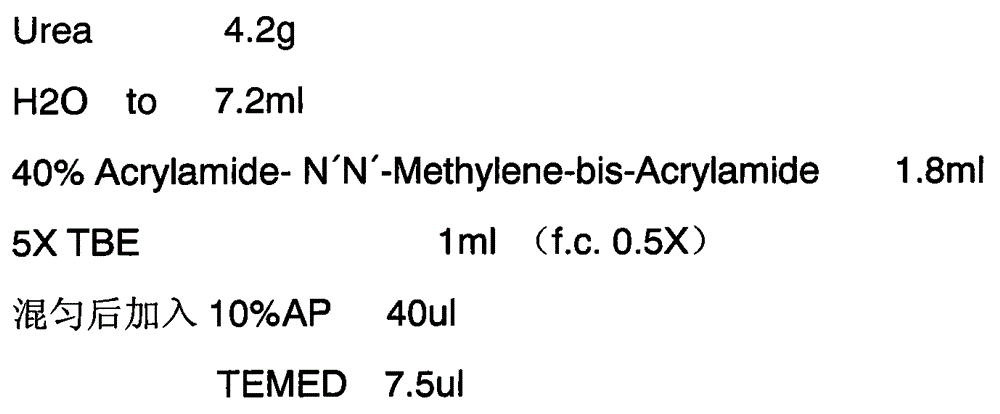
[0025] Before further describing the specific embodiments of the present invention, it should be understood that the protection scope of the present invention is not limited to the following specific specific embodiments, and it should also be understood that the terms used in the embodiments of the present invention are for describing specific specific implementations scheme, not to limit the scope of protection of the present invention. The following describes the construction process of the CLIP-seq library in Arabidopsis by taking the newly identified homologous protein hnRNP A1-like protein 1 (HLP1) of hnRNP A1 in Arabidopsis as an example.
[0026] 1. Ultraviolet Crosslinking
[0027] Take a 150mm cell culture dish, cut two filter papers of appropriate size, spread one in the dish, and pour 150ml of ice-cold PBS into it. Collect the seedlings grown on MS medium for 8-14 days (the specific growth period needs to be determined according to the expression period of the spe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com