Sequencing joint and preparation method and application thereof in ultra-low frequency mutation detection
A sequencing linker and ultra-low frequency technology, which is applied in biochemical equipment and methods, DNA preparation, microbial measurement/inspection, etc., can solve the problem of insufficient detection accuracy and achieve the effect of improving detection accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0036] In another typical embodiment of the present application, a method for preparing a sequencing linker is also provided, the preparation method comprising: designing two single-stranded sequences of the sequencing linker, each single-stranded sequence containing an error prompt sequence, and The error prompt sequence is located on the side of the single-stranded sequence close to the target fragment to be connected, and the error prompt sequence is a sequence with a known base sequence; two single-stranded sequences are synthesized respectively; the two synthetic single-stranded sequences are specifically identified Anneal to form double-stranded sequencing adapters.
[0037] The synthesis in the above preparation method refers to chemical synthesis, which is usually synthesized by a company that provides chemical synthesis services. Rather than biosynthesis, that is, it does not refer to synthesis by using one strand as a template and the other through replication and ex...
Embodiment 1
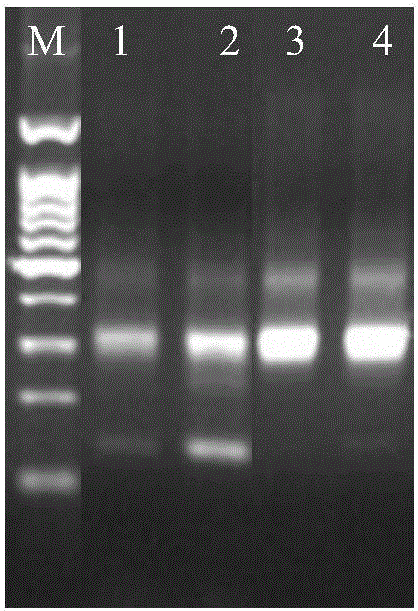
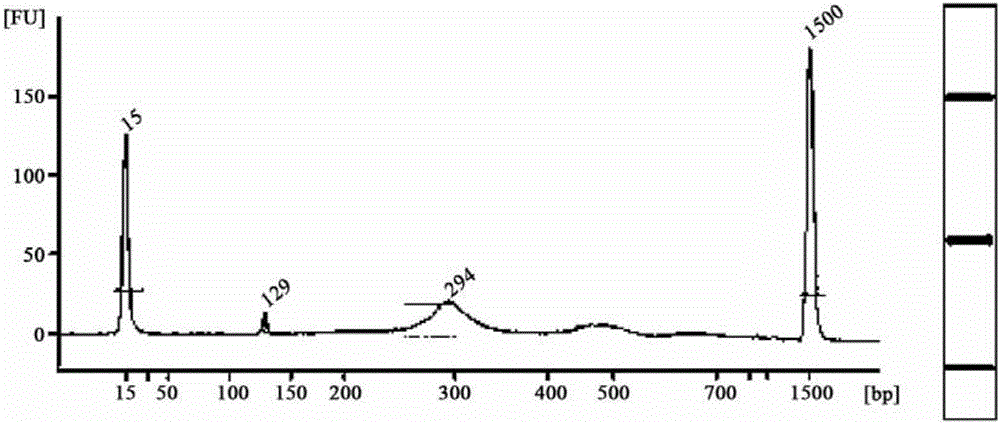
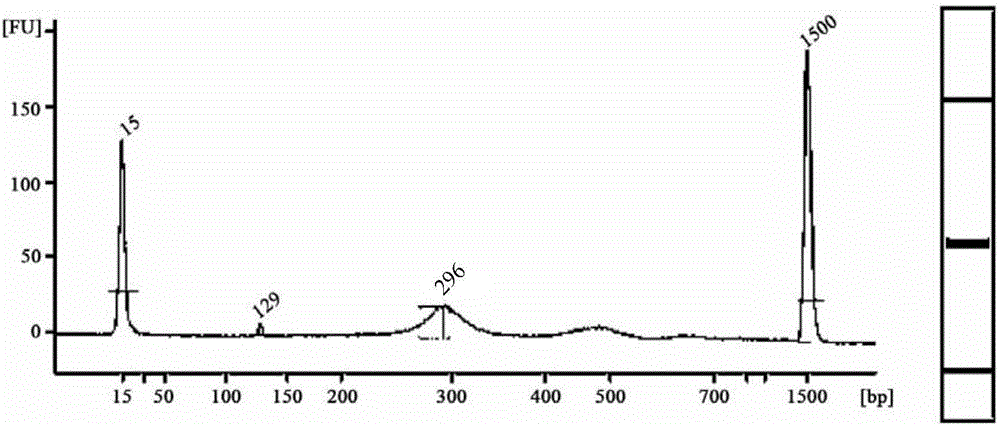
[0075] 2. Embodiment 1 joint (2) manufacture method
[0076] (1) Linker sequence synthesis
[0077] ERROR-top: ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNNT (SEQ ID NO: 5);
[0078] ERROR-bot: P*NNNNNNNAGATCGGAAGAGCACACGTCTGAACTCCAGTCAC (SEQ ID NO: 6: ). Remarks: P* is phosphorylation modification
[0079] In Example 1, 100 kinds of error prompt sequences were specifically set for N parts, and the ERROR-bot tag sequence was reversely complementary to the corresponding ERROR-top error prompt sequence. Among them, Table 4 shows 100 kinds of ERROR-top sequences. ERROR-top and the corresponding ERROR-bot are synthesized separately during synthesis. Among them, among the 7 bases shown in bold, except that the T at the end is for connecting with the protruding A at the 3' end of the target fragment, the remaining 6 bases are error prompt sequences.
[0080] Table 4:
[0081]
[0082]
[0083]
[0084]
[0085] (2) Dilute the powder of each primer with nuclease-free wat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com