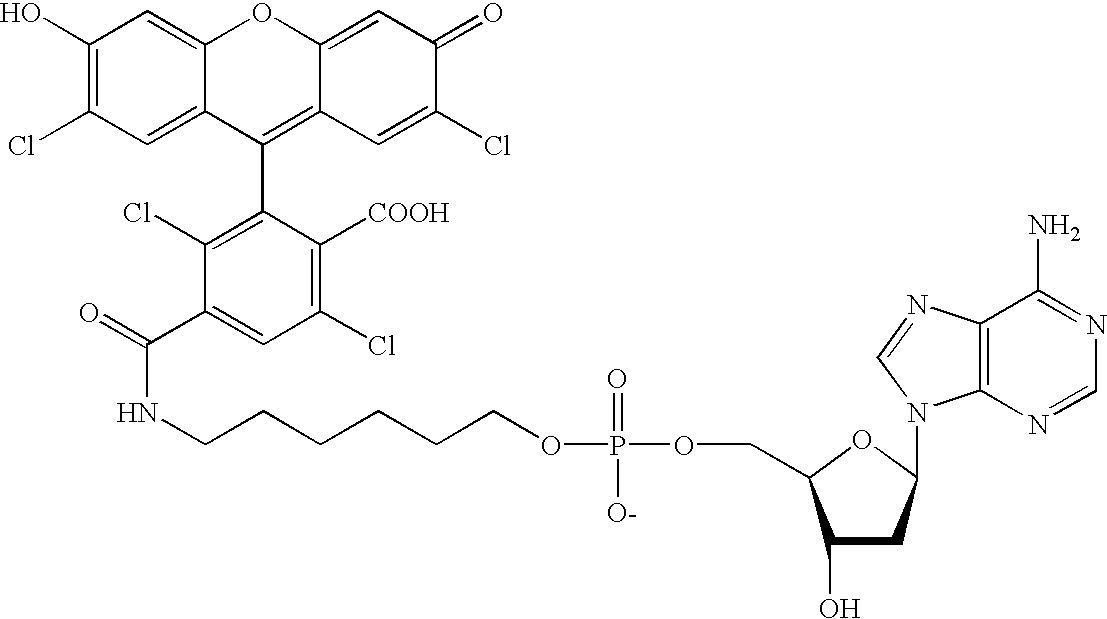
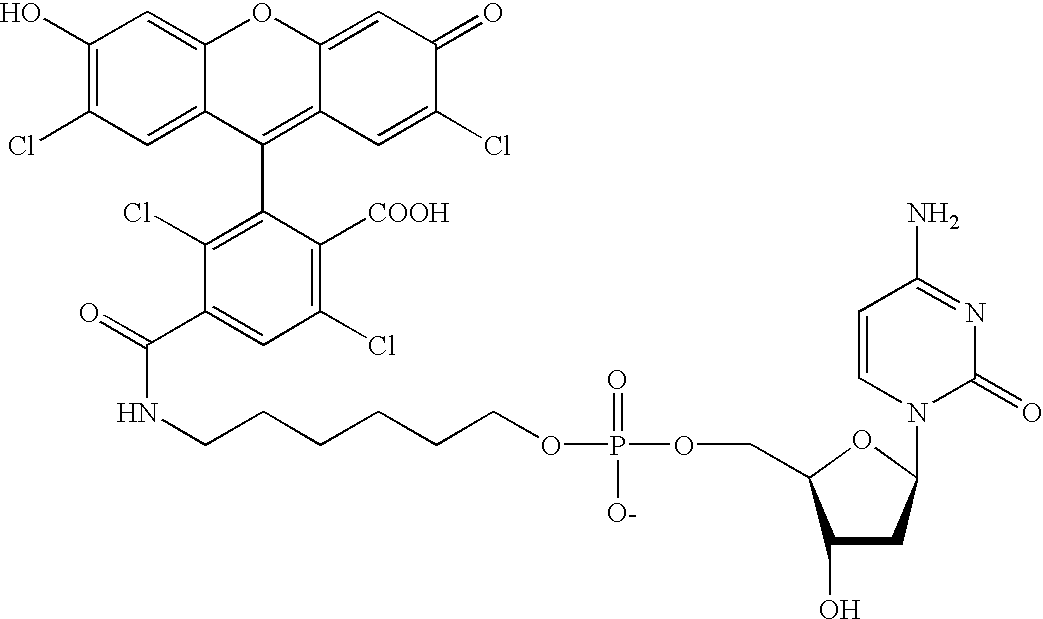
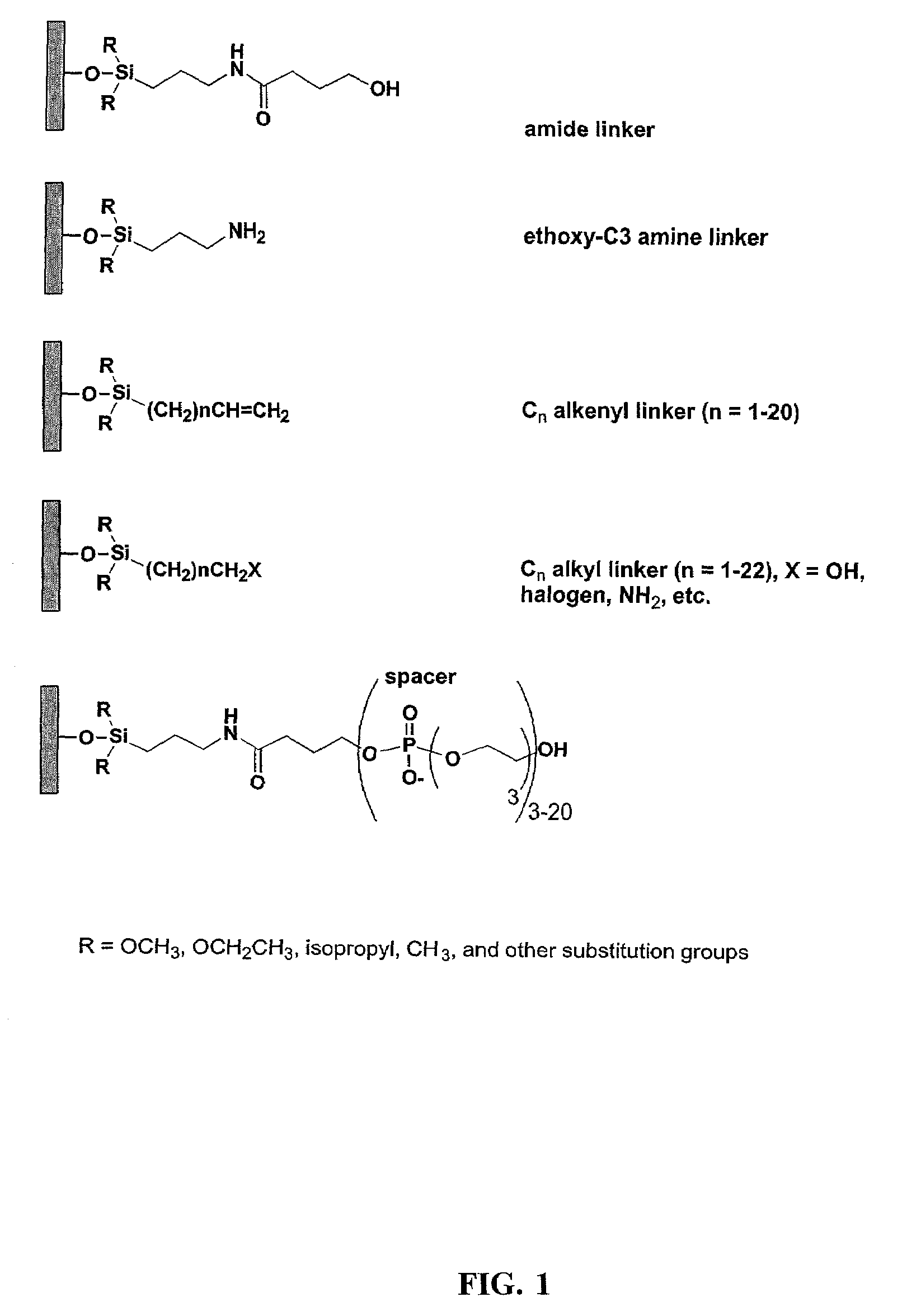
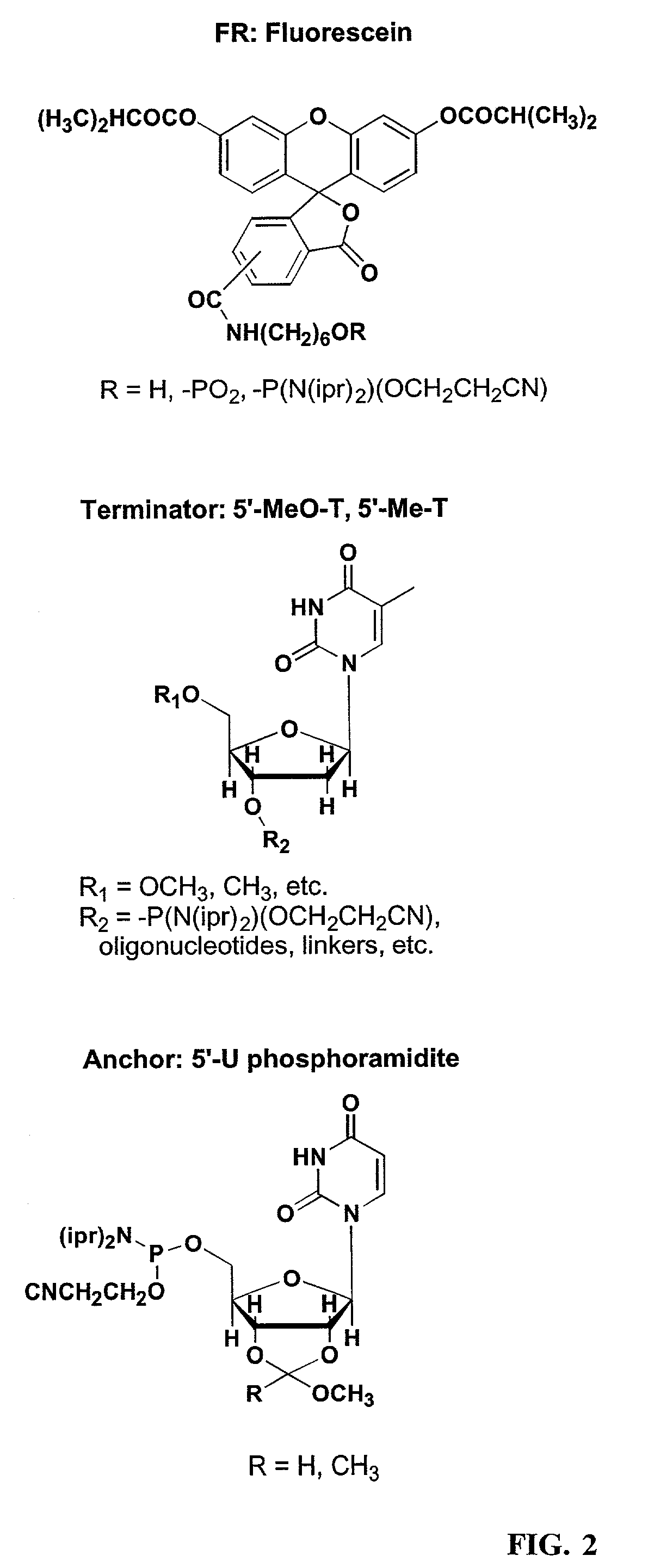
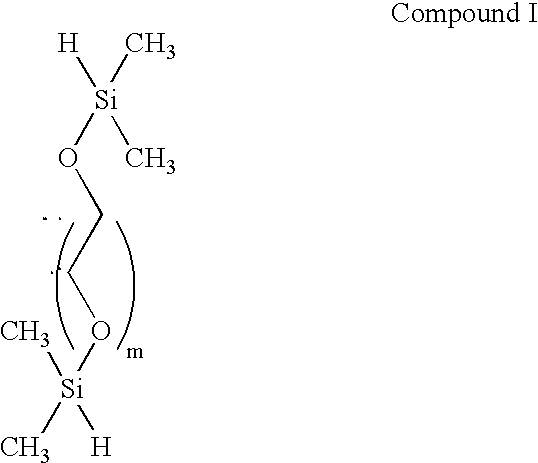
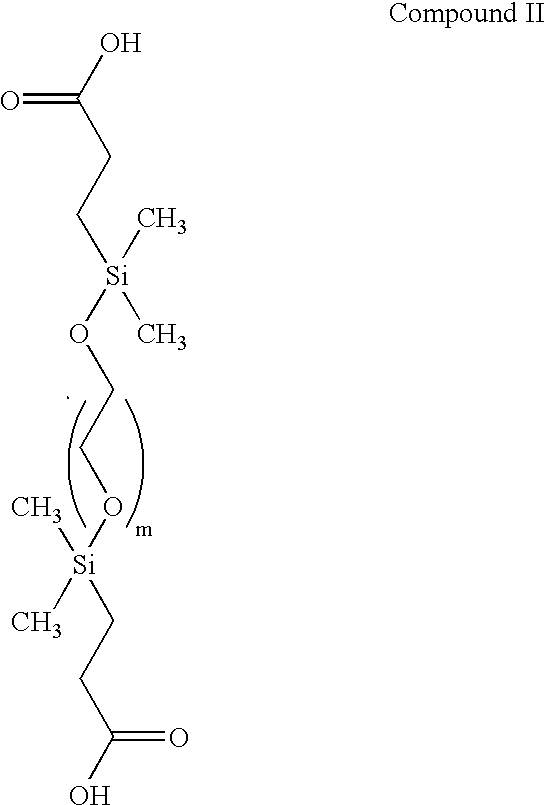
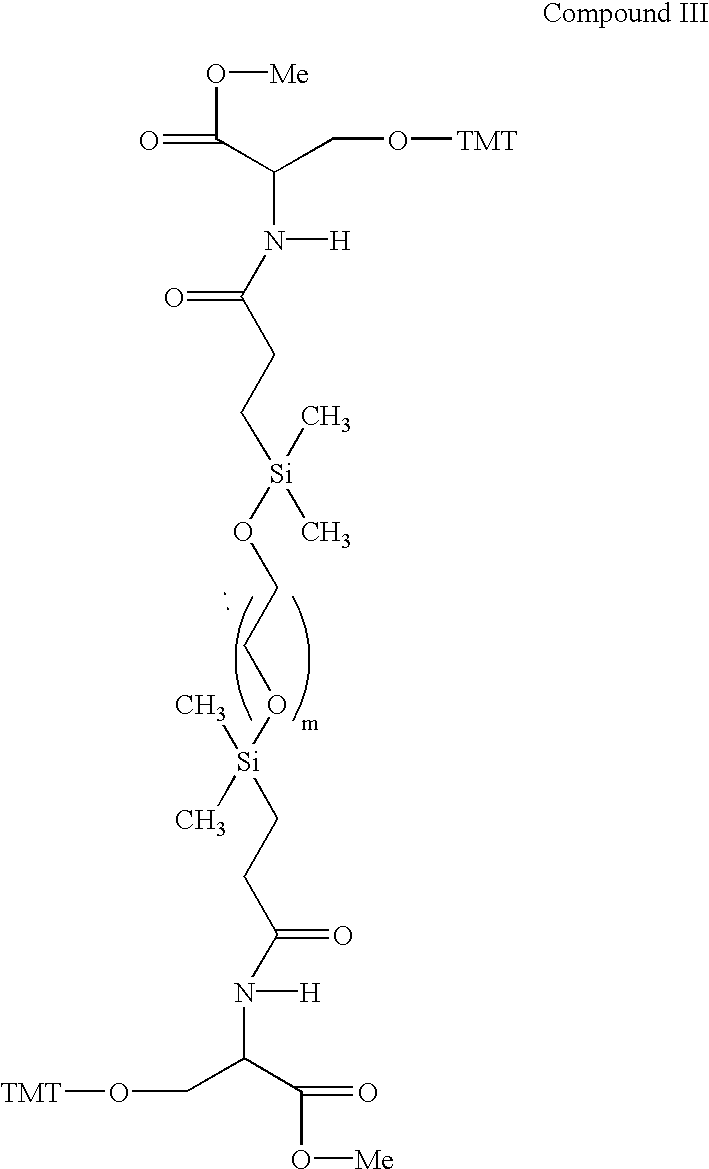
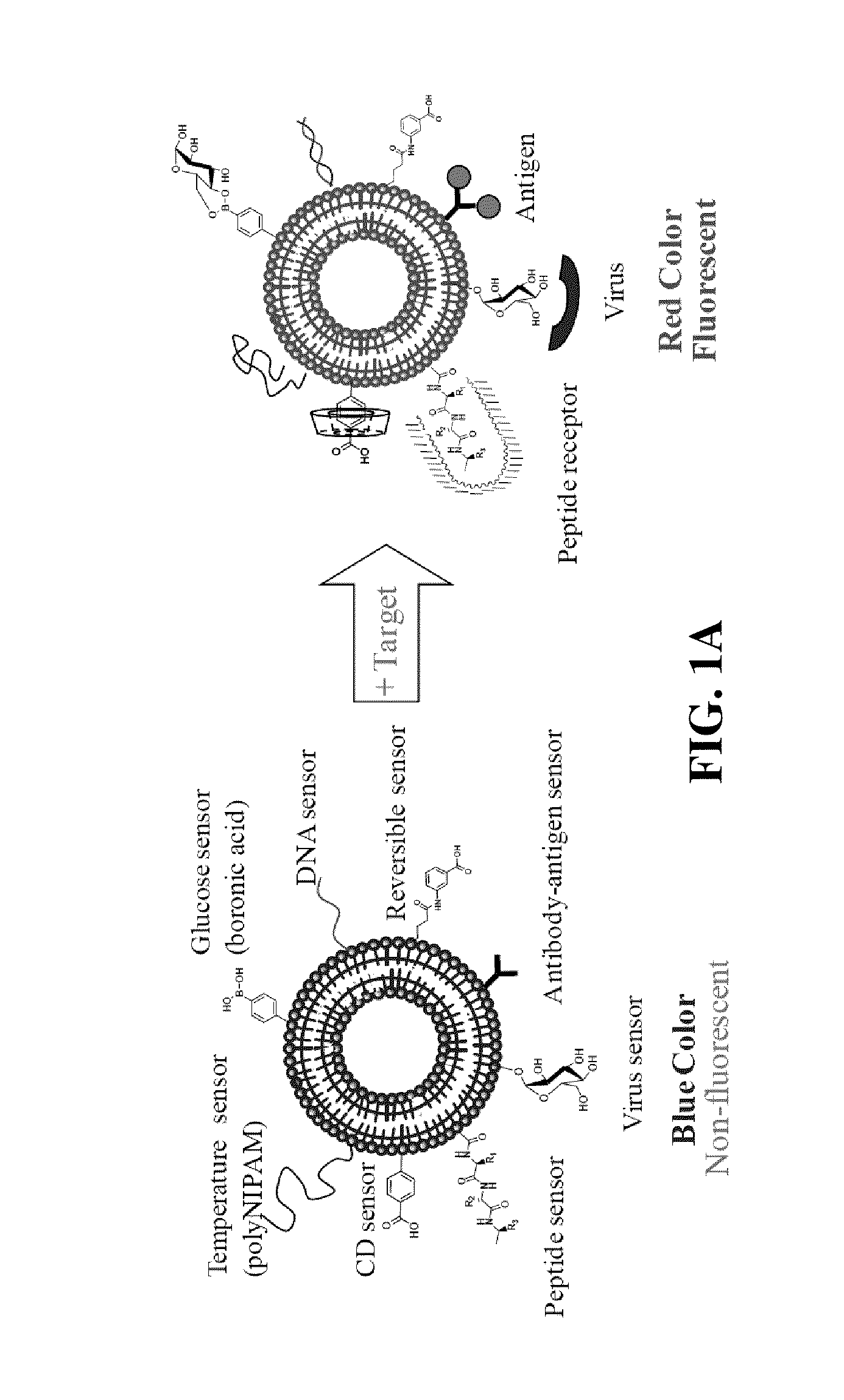
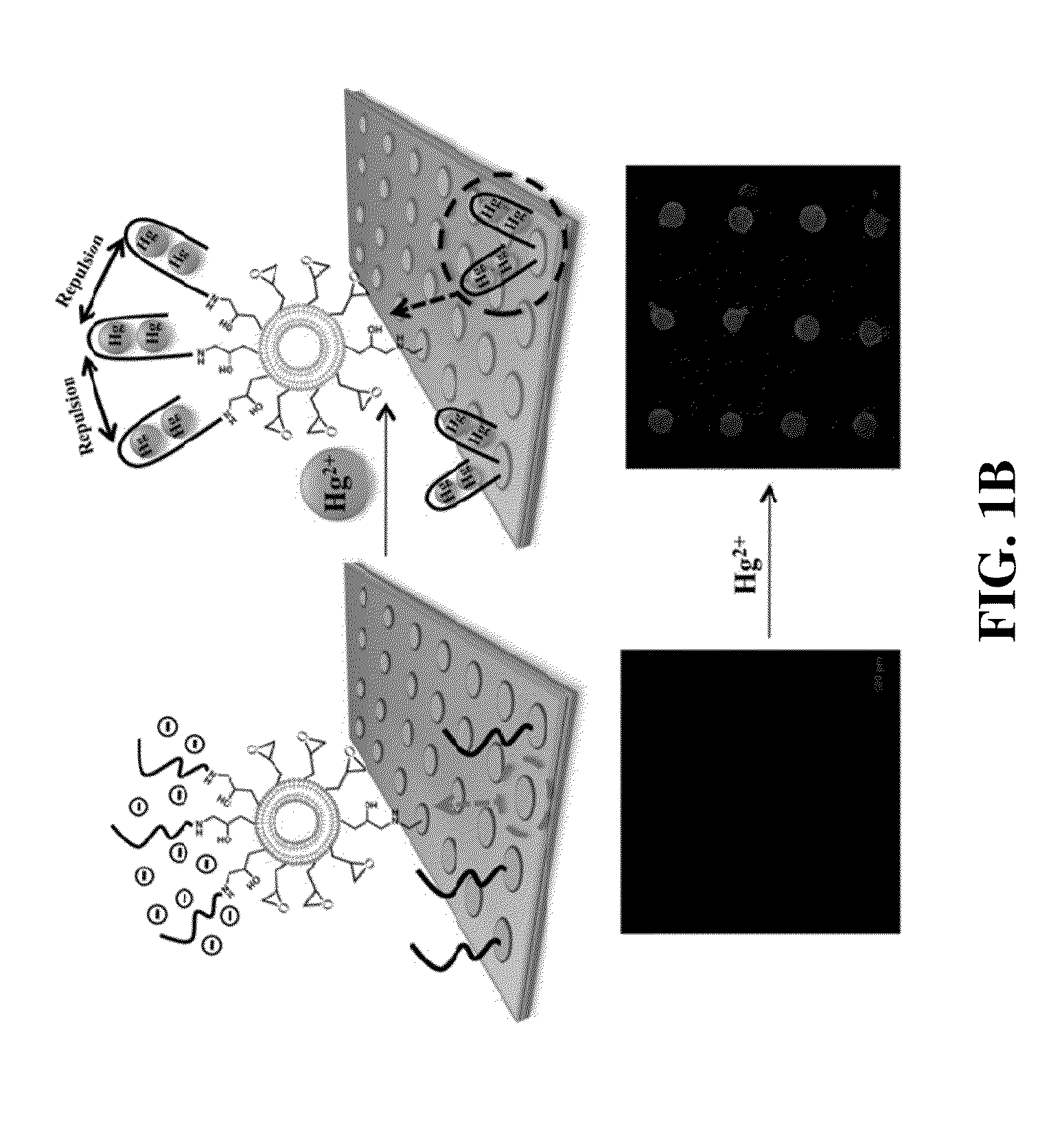
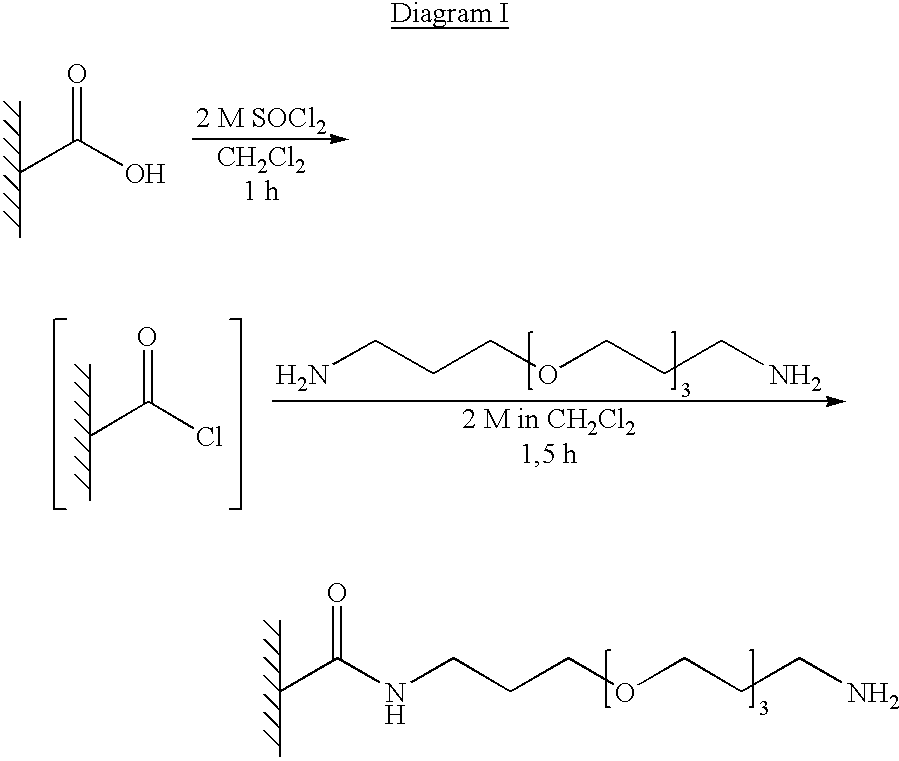
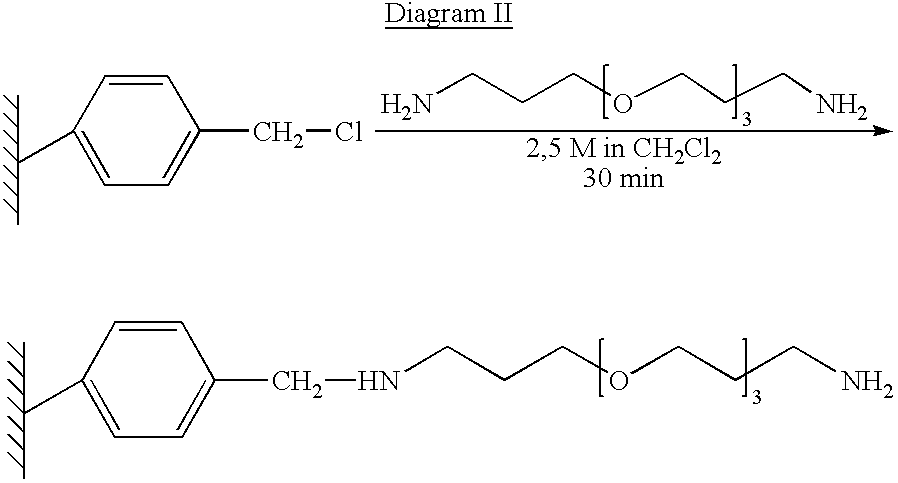
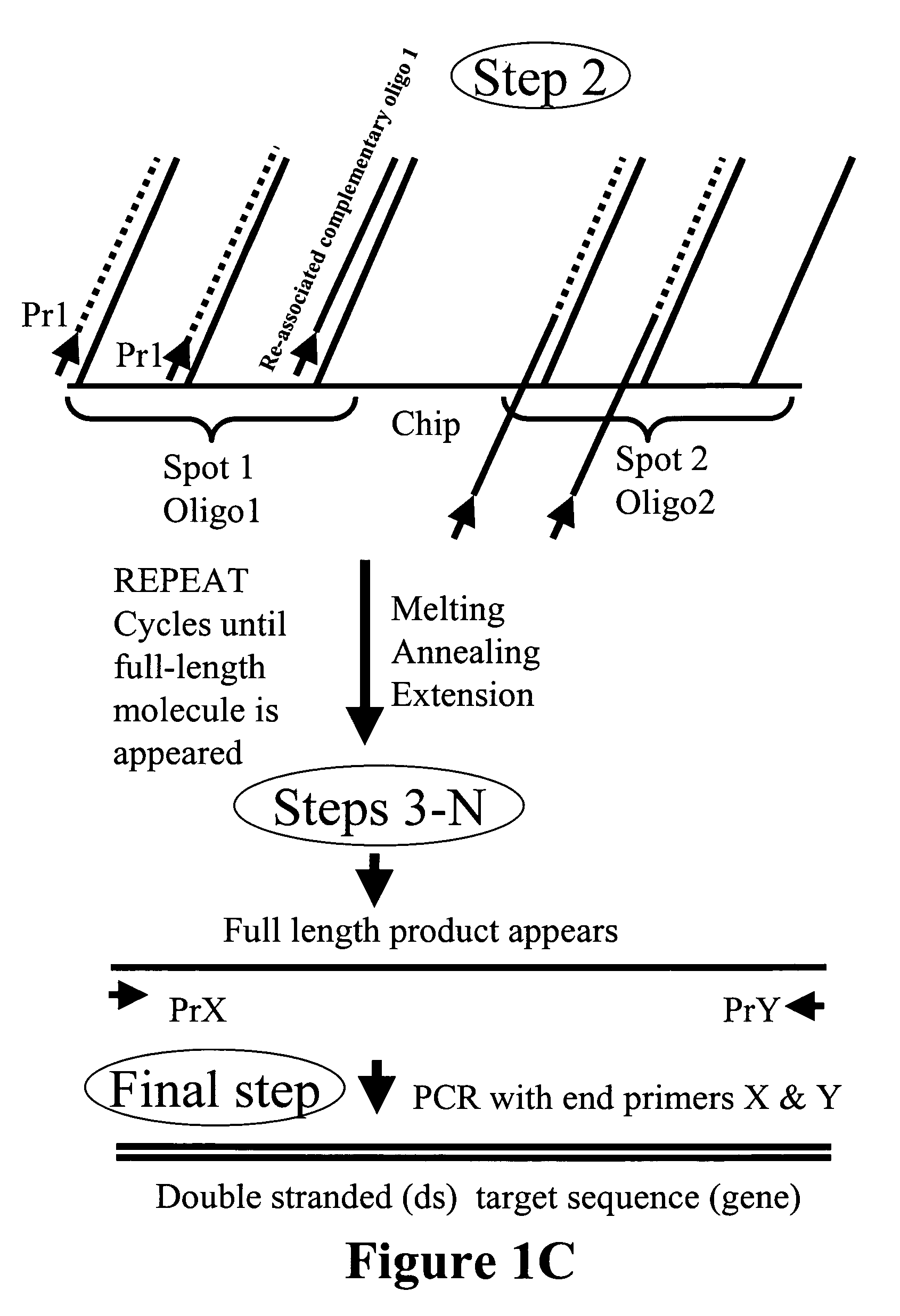
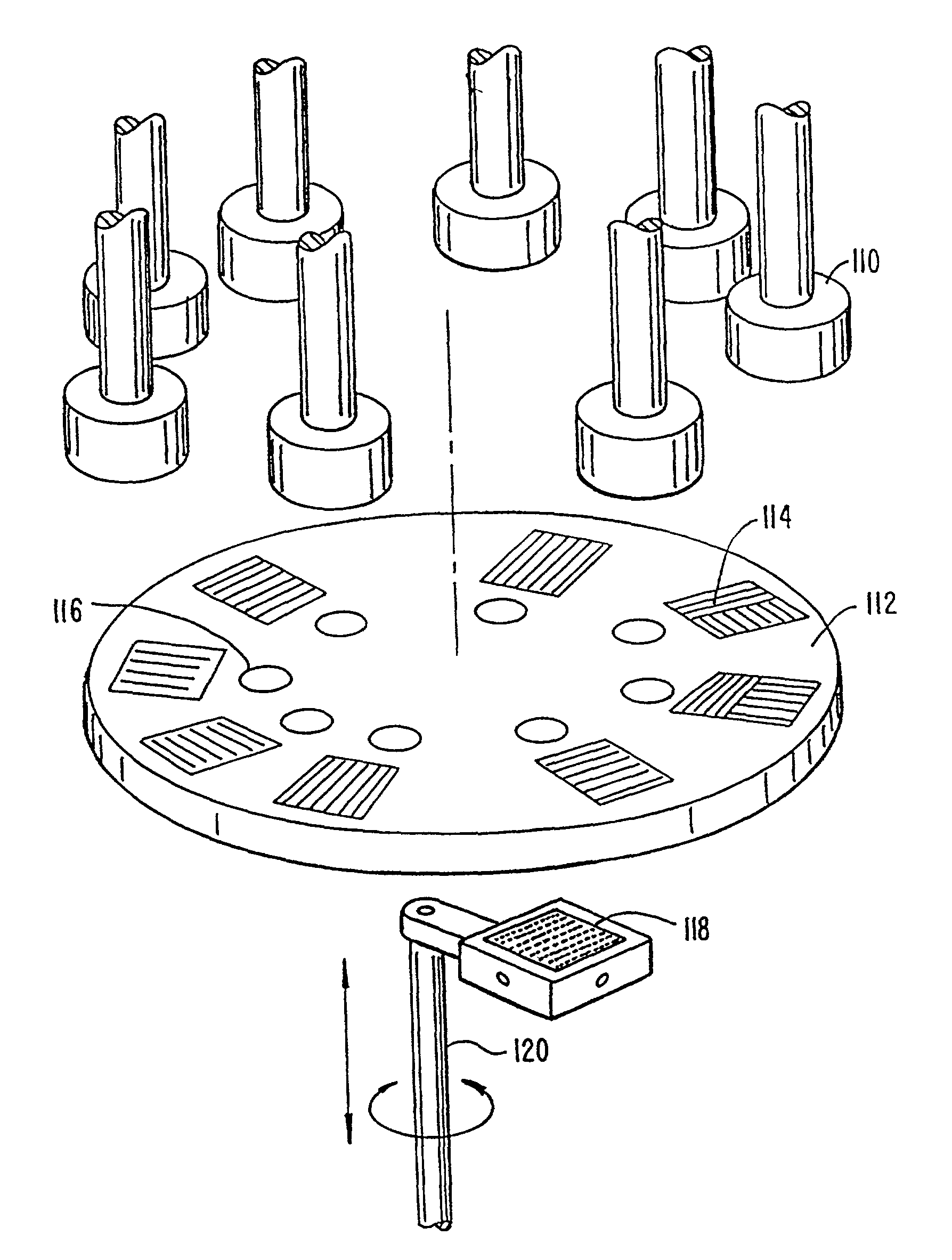
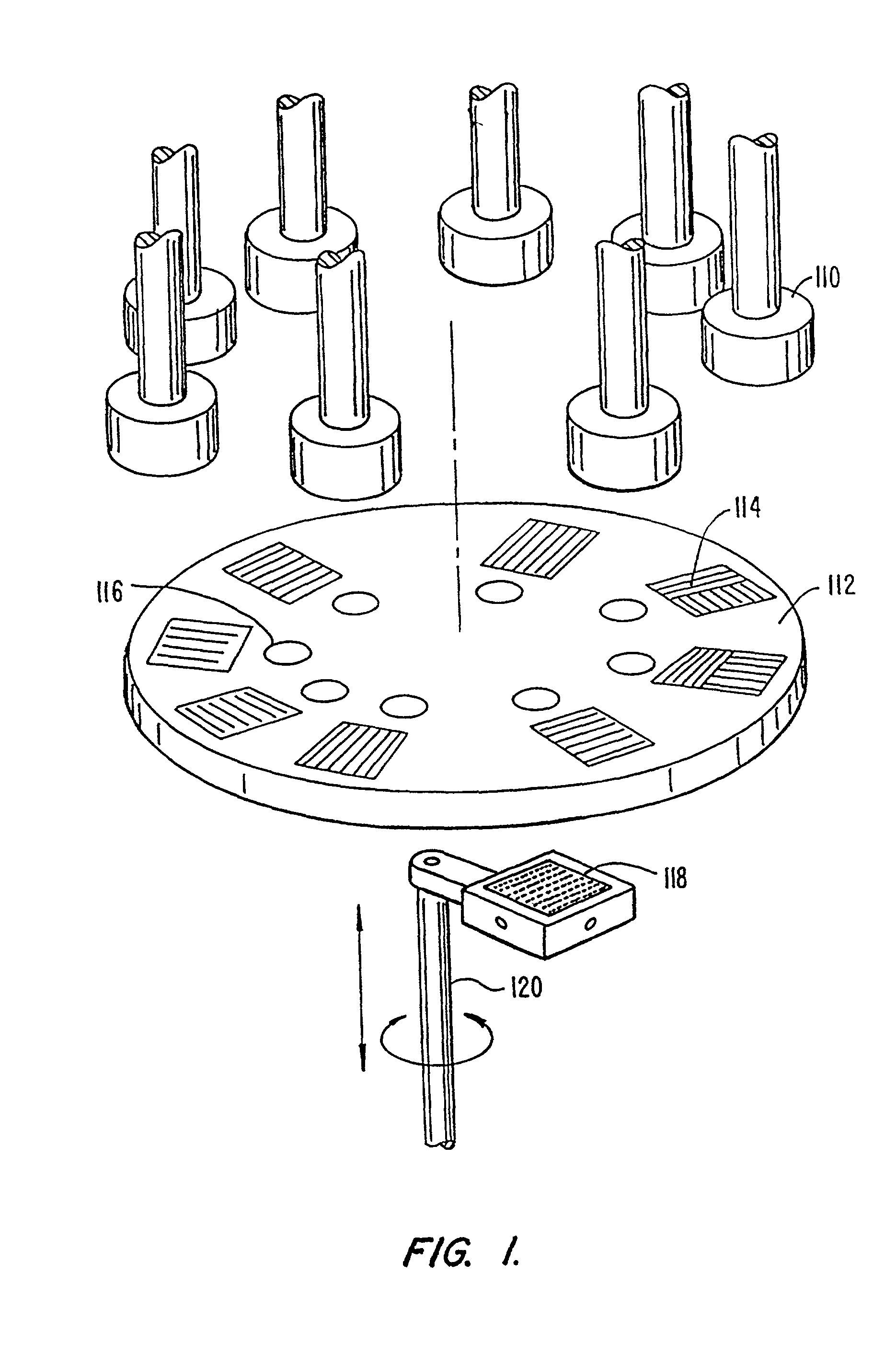
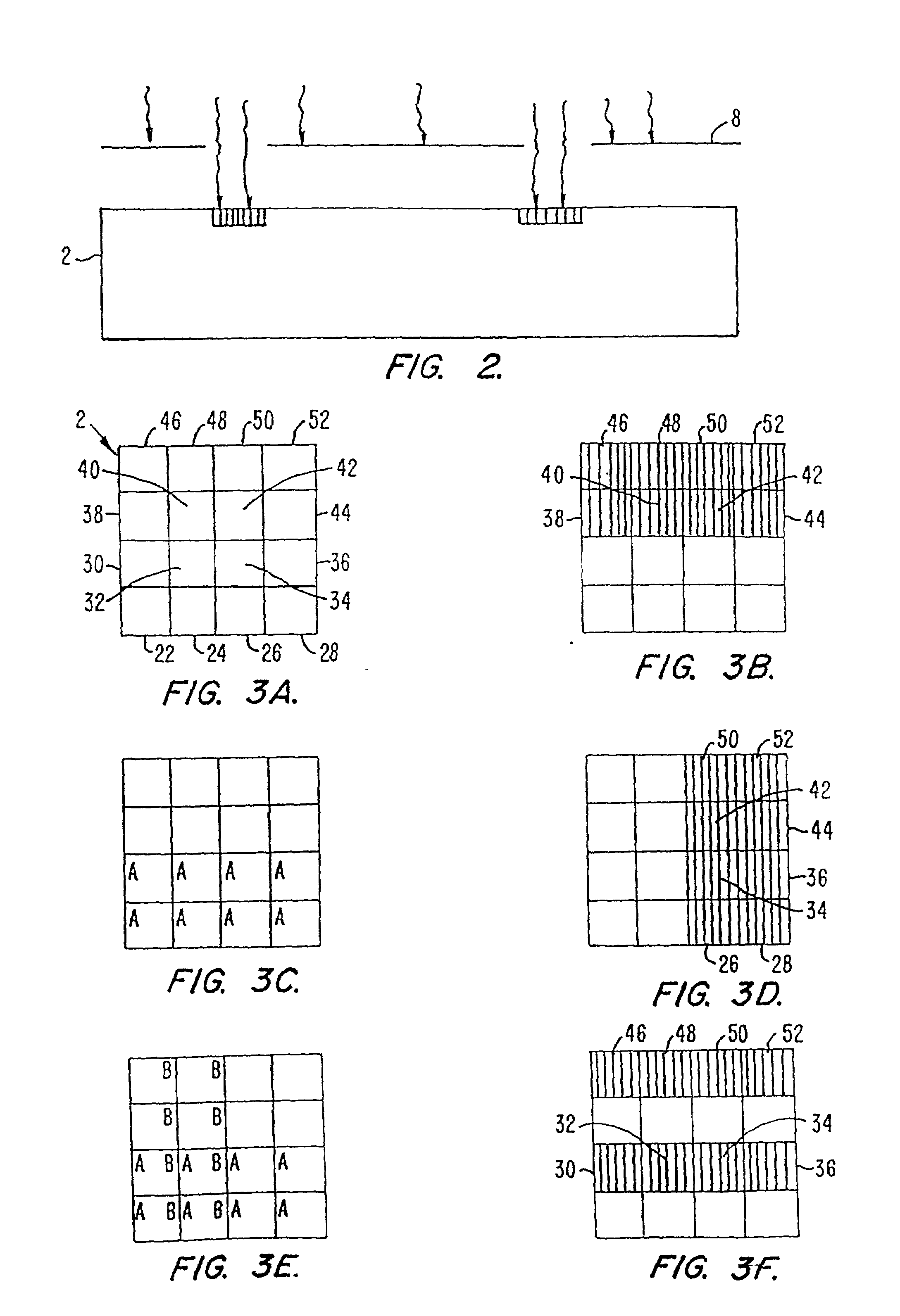
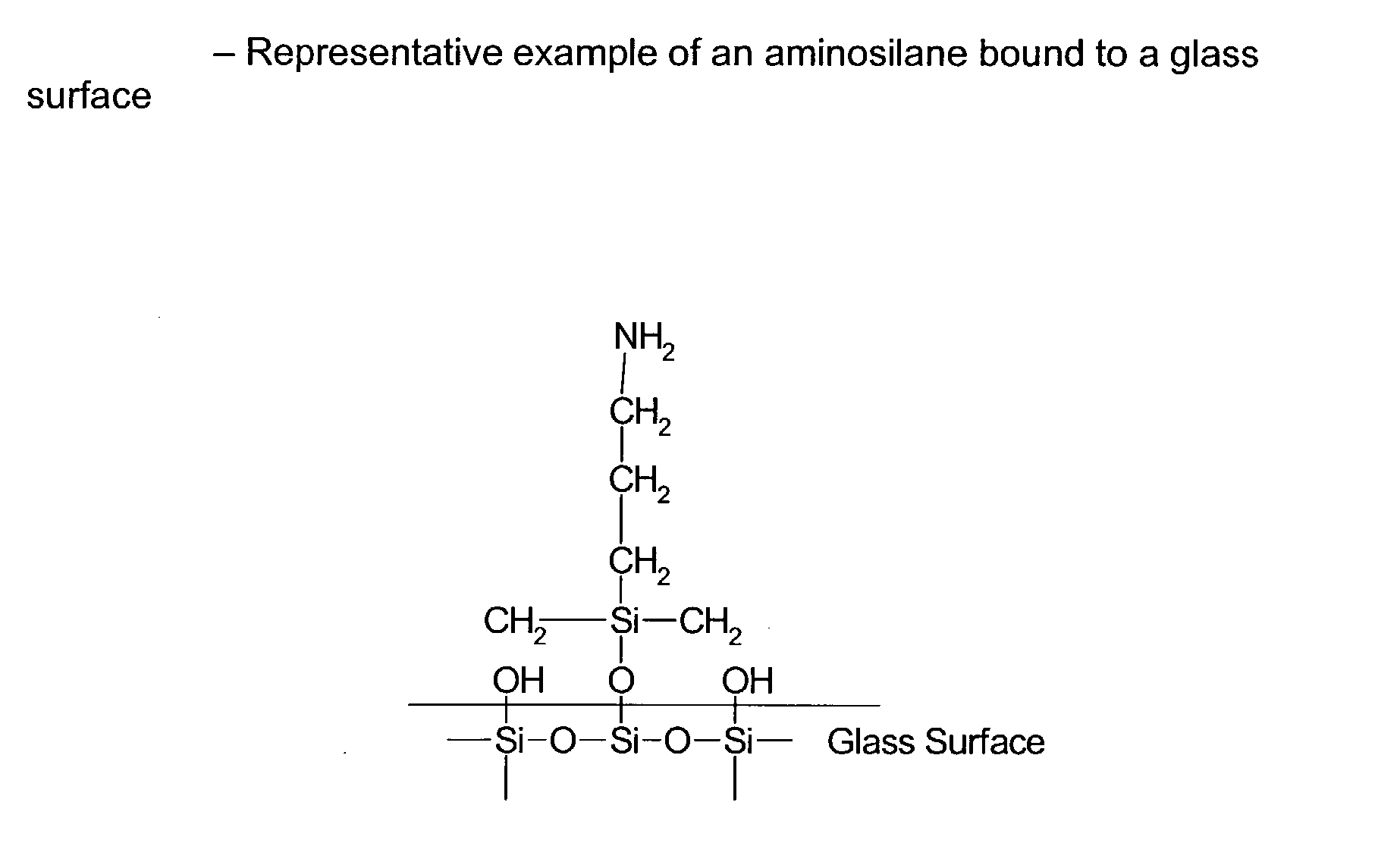
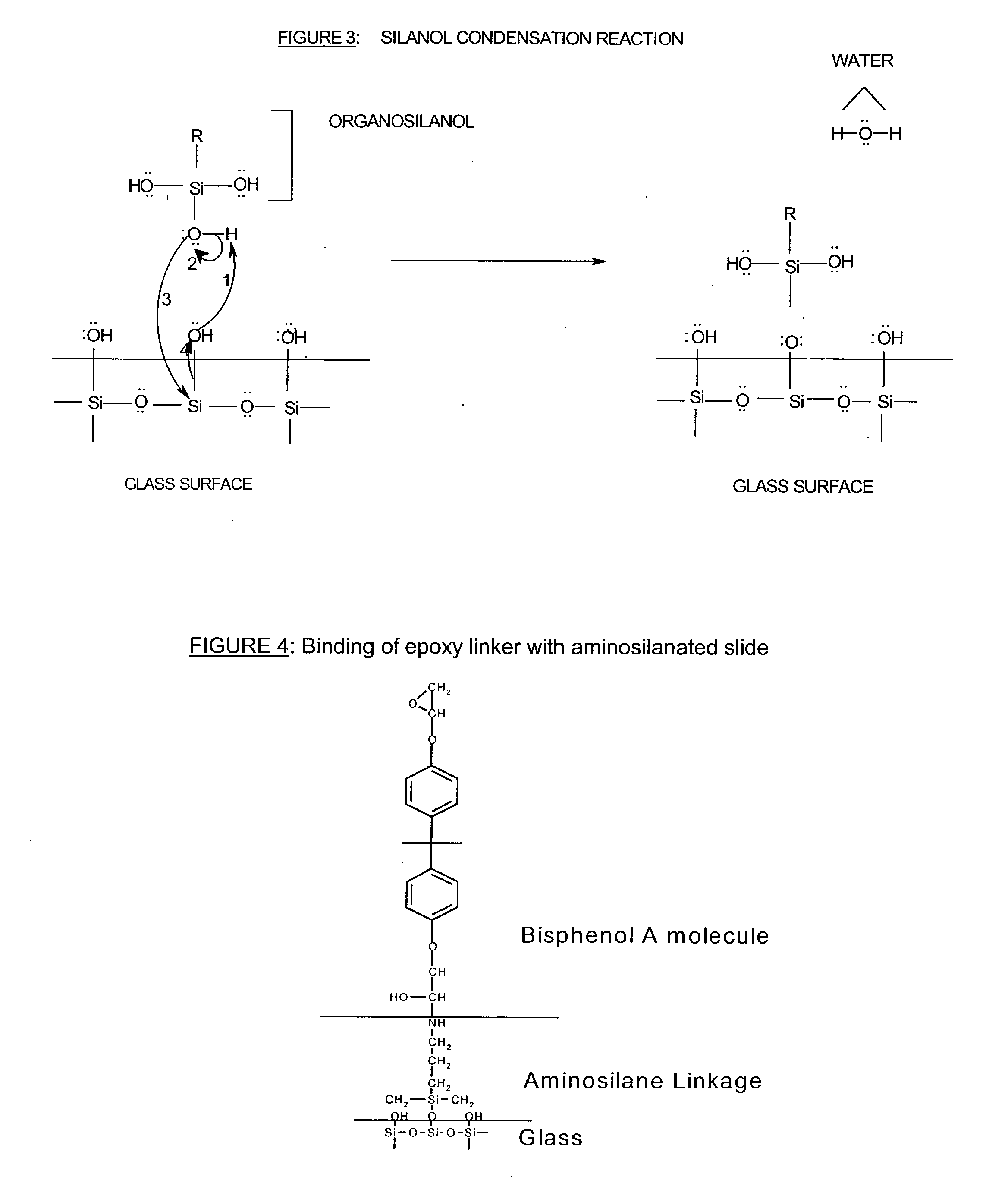
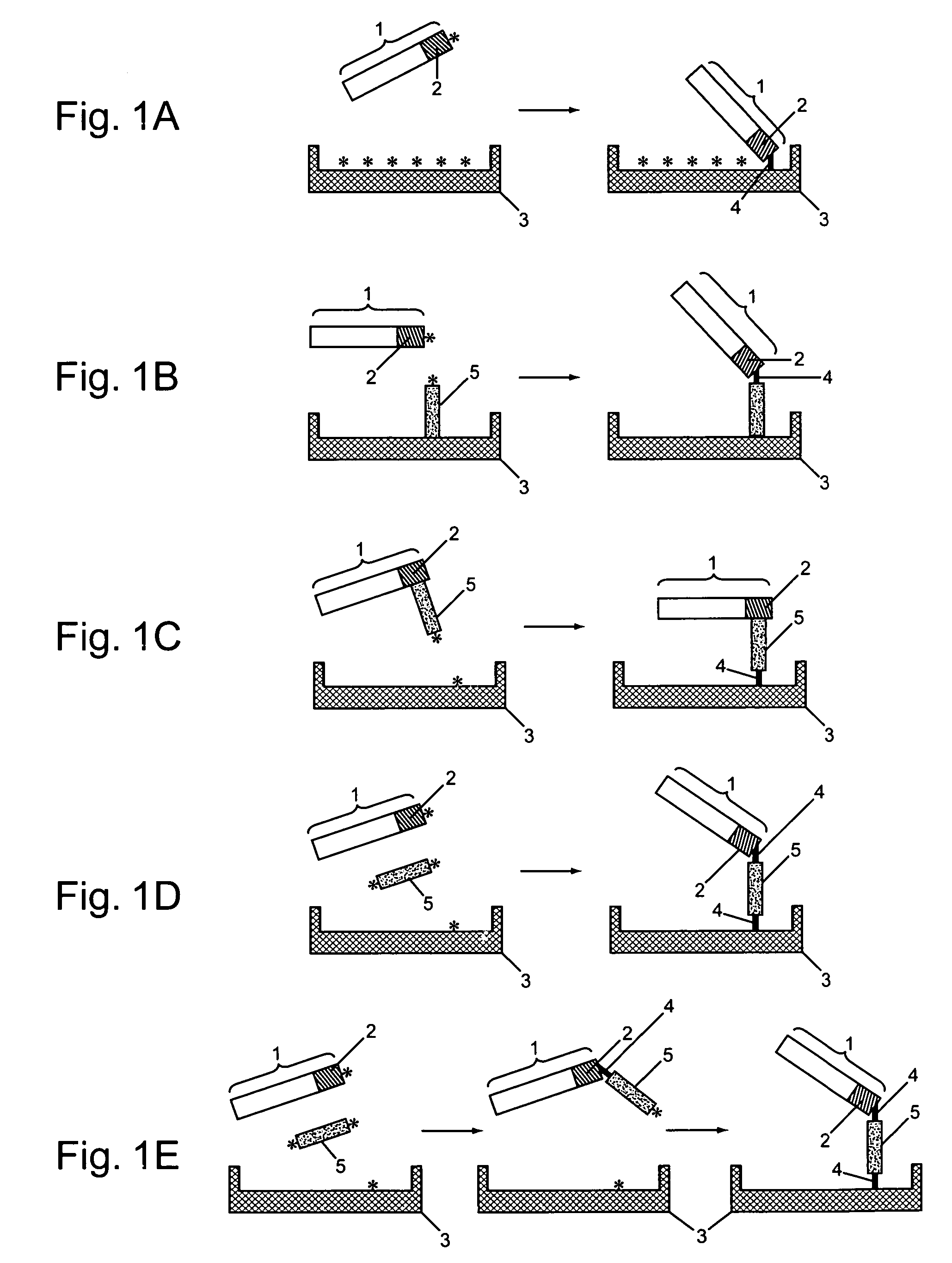
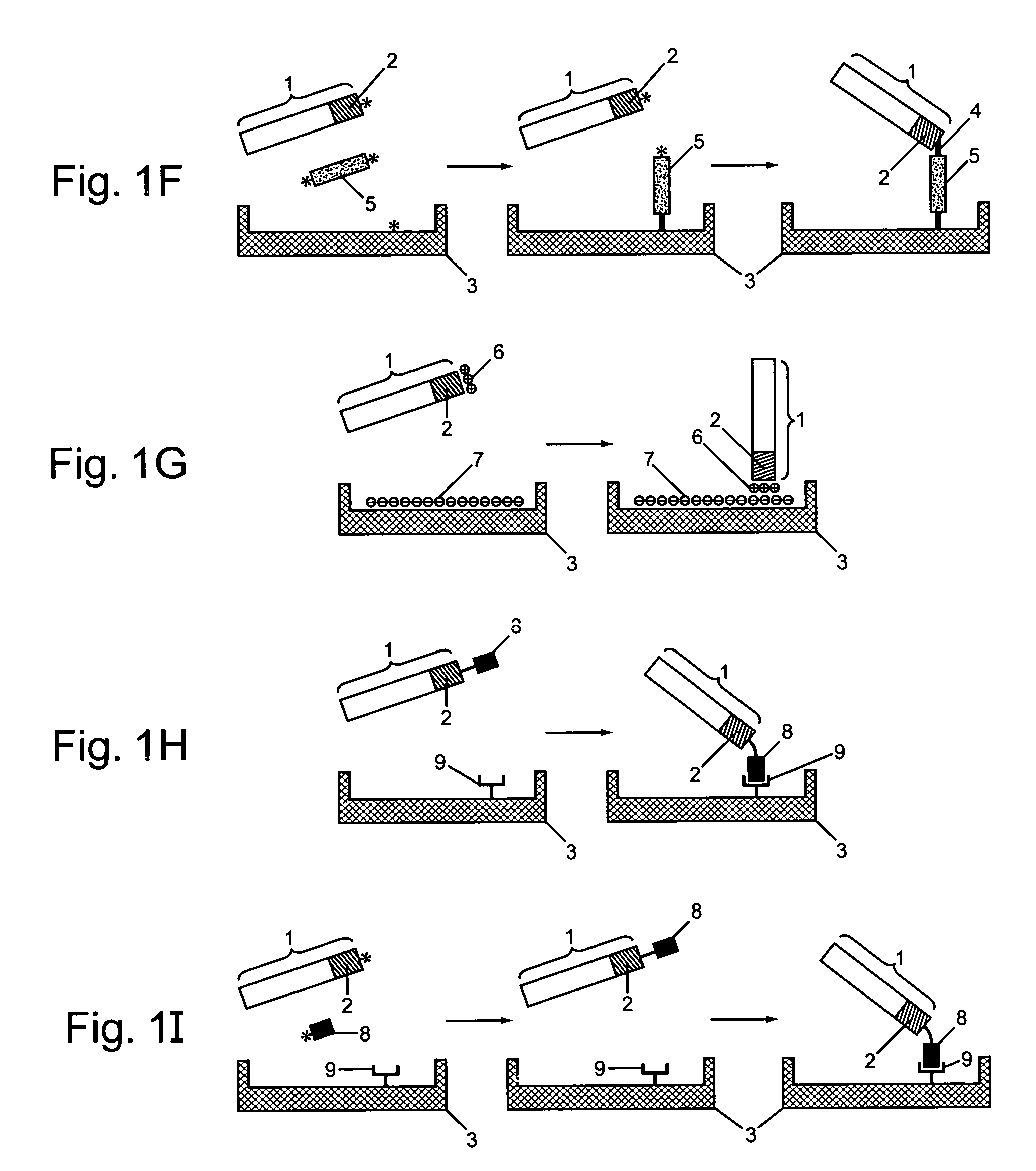
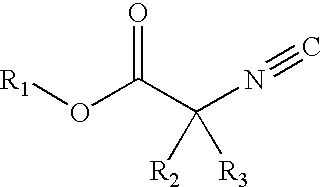
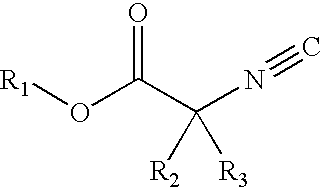
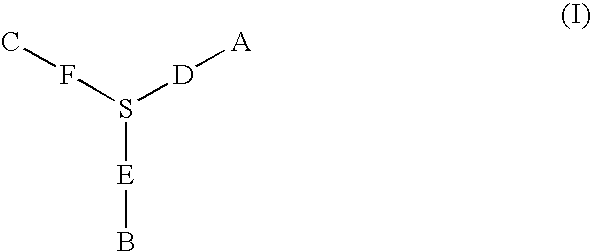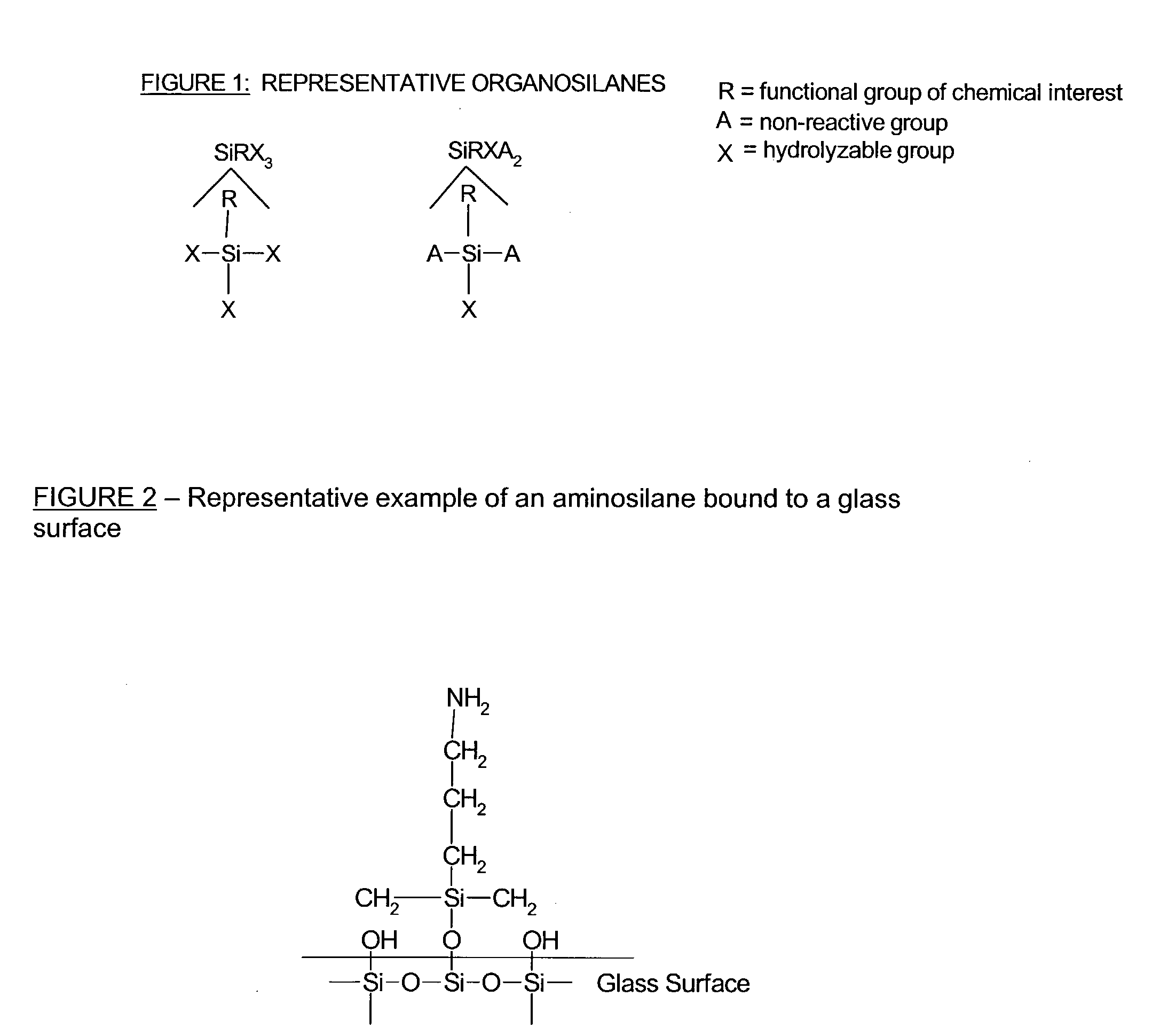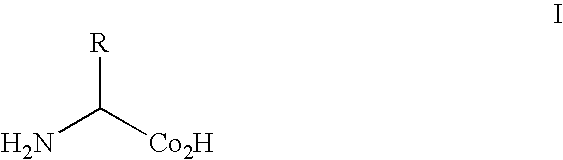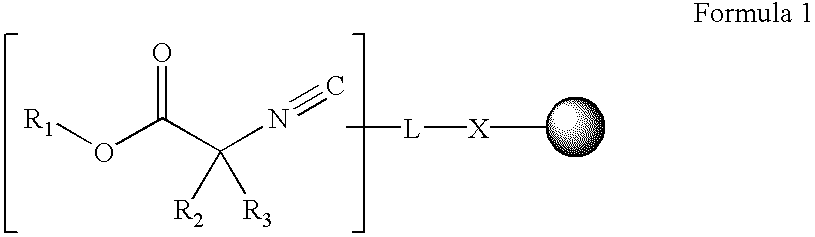Patents
Literature
156results about "Library linkers" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
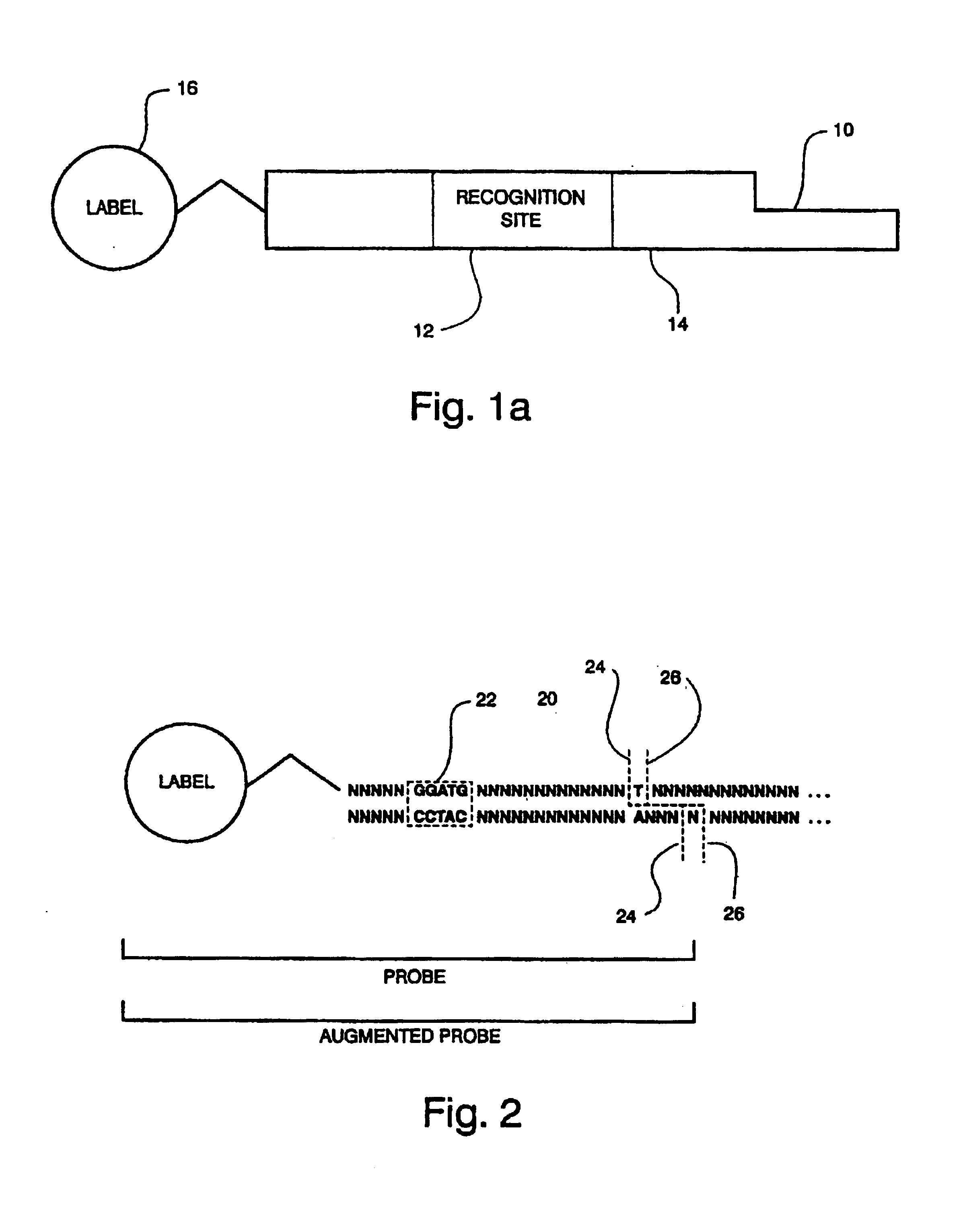
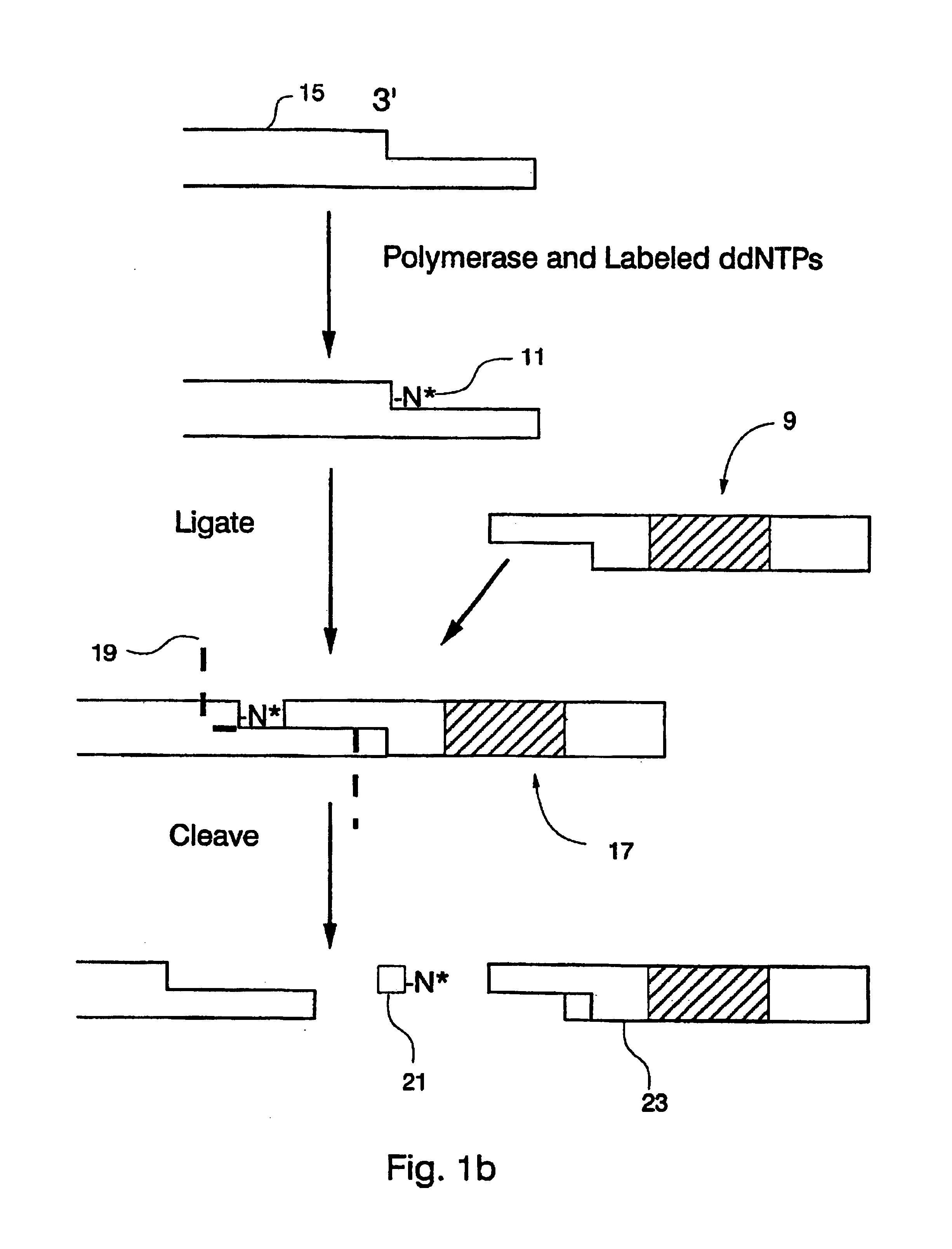
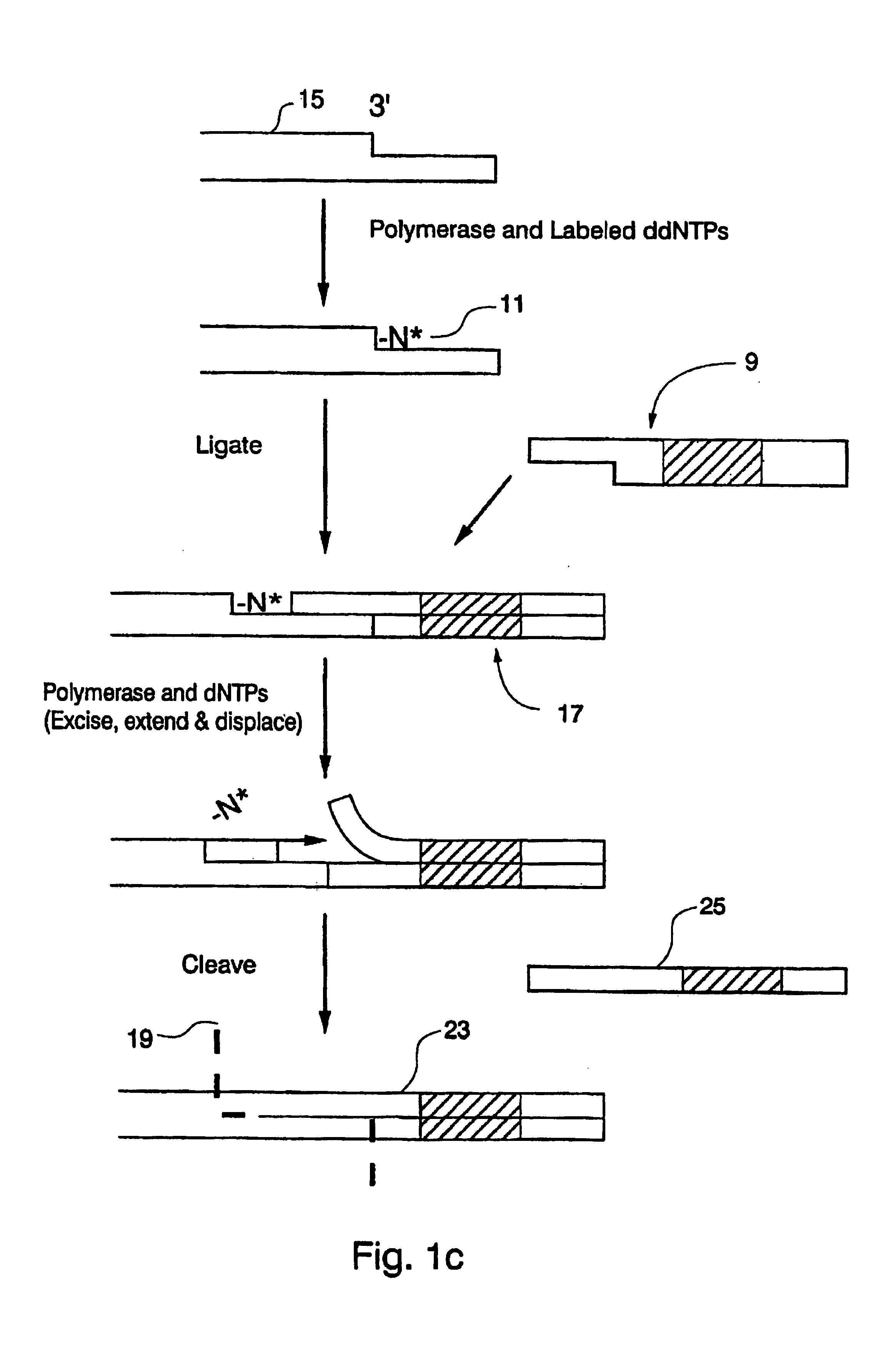
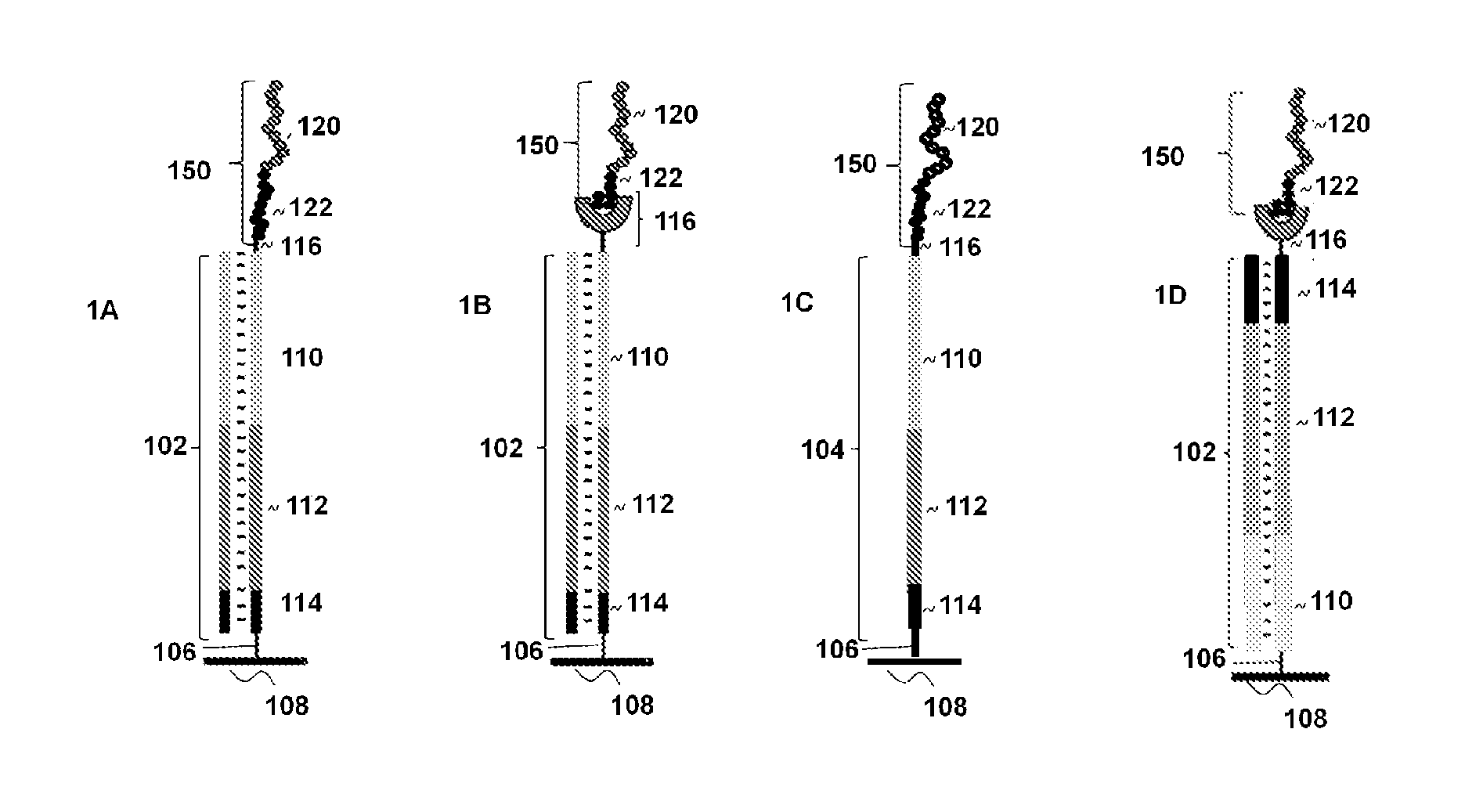
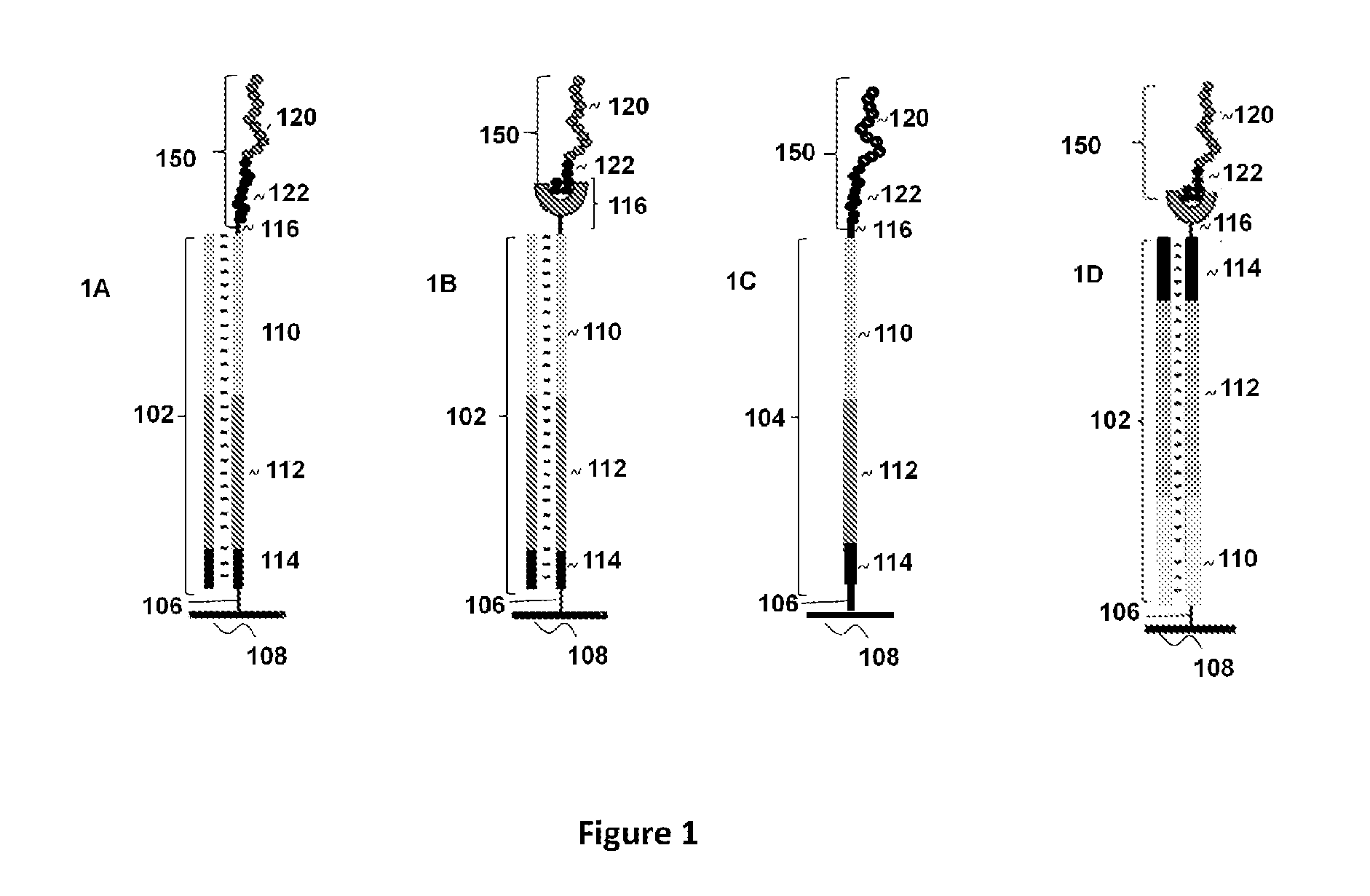
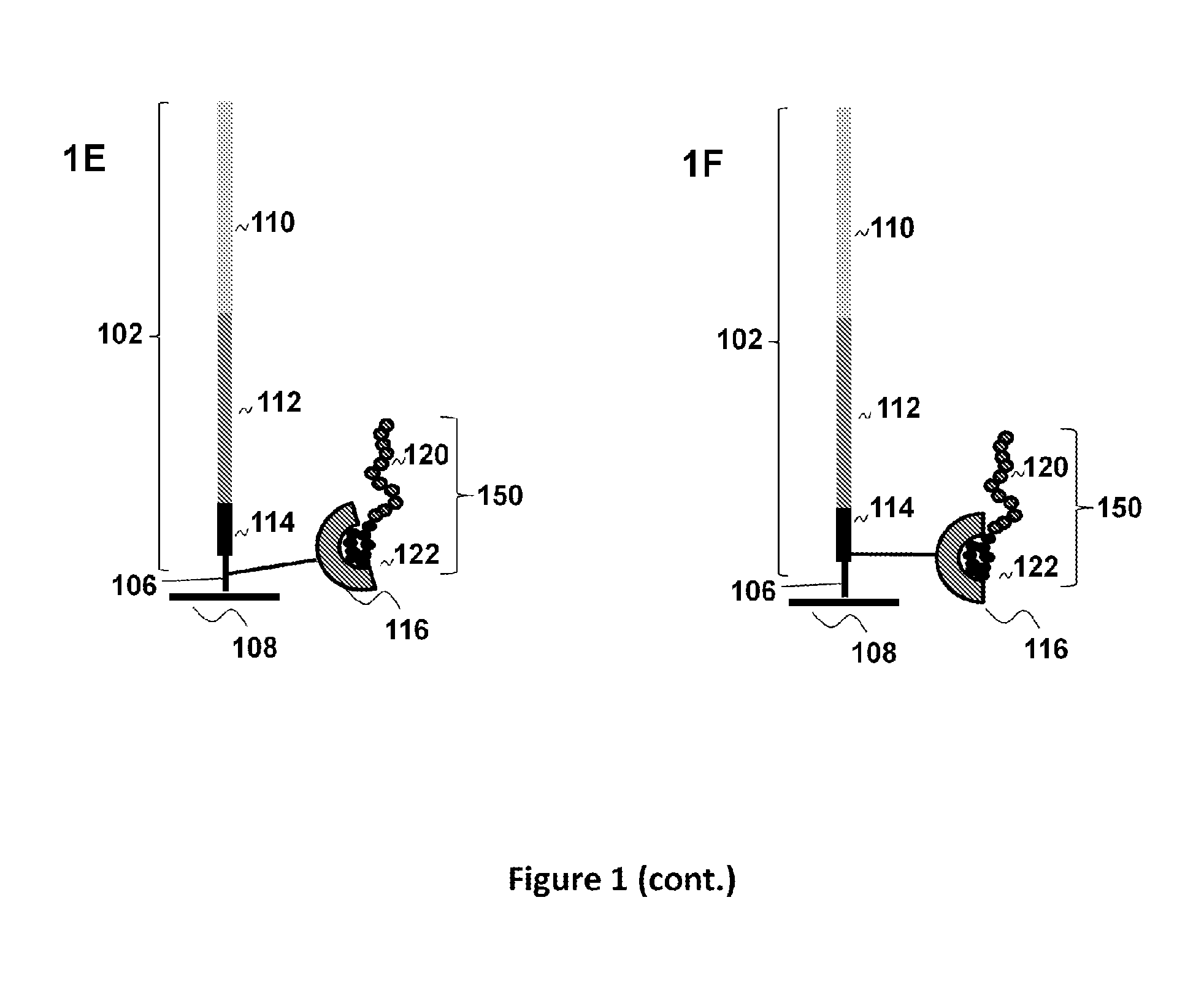
Optical disk-based assay devices and methods
InactiveUS6342349B1Strong specificityReduce nonspecific bindingSequential/parallel process reactionsSugar derivativesAnalyteLaser light
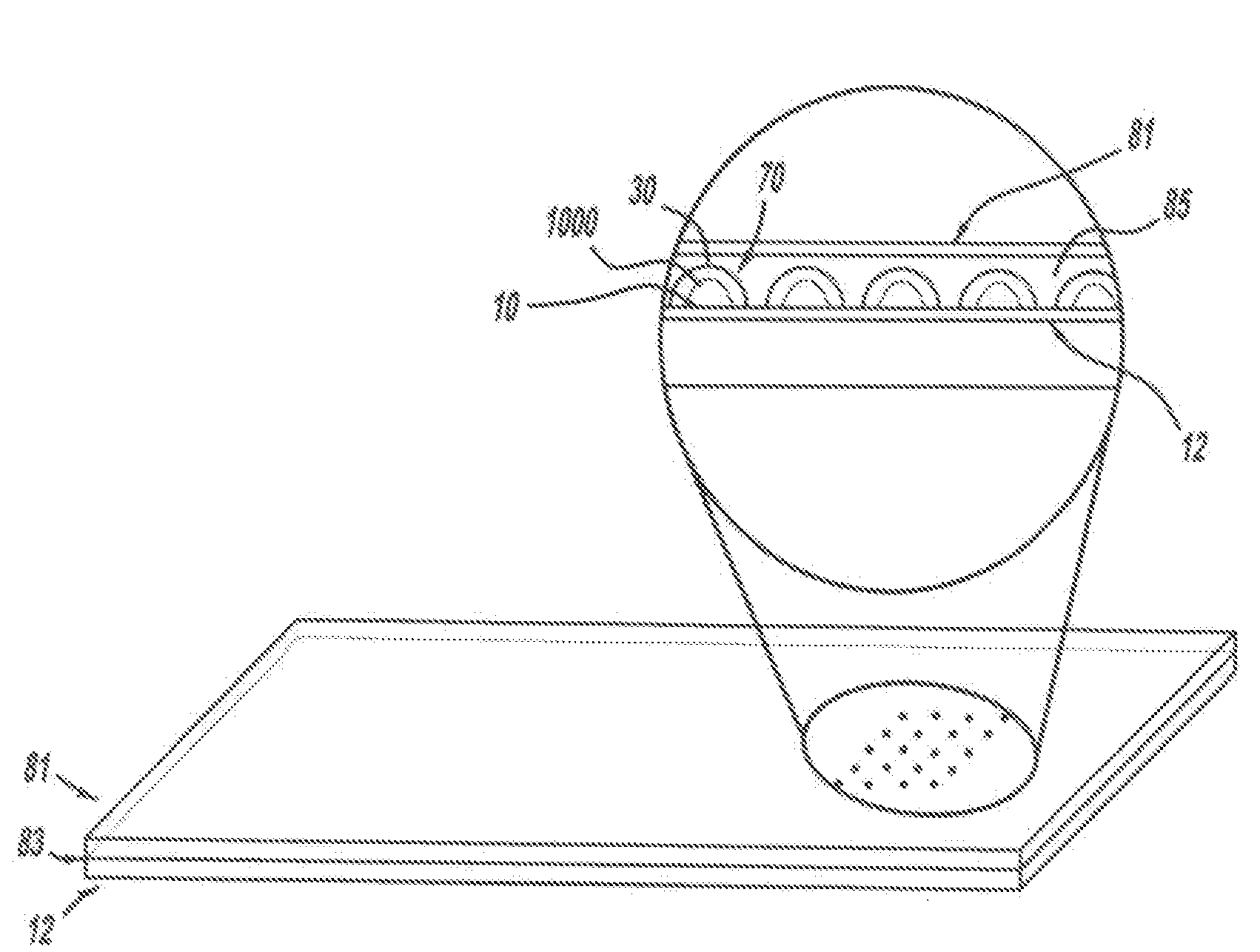
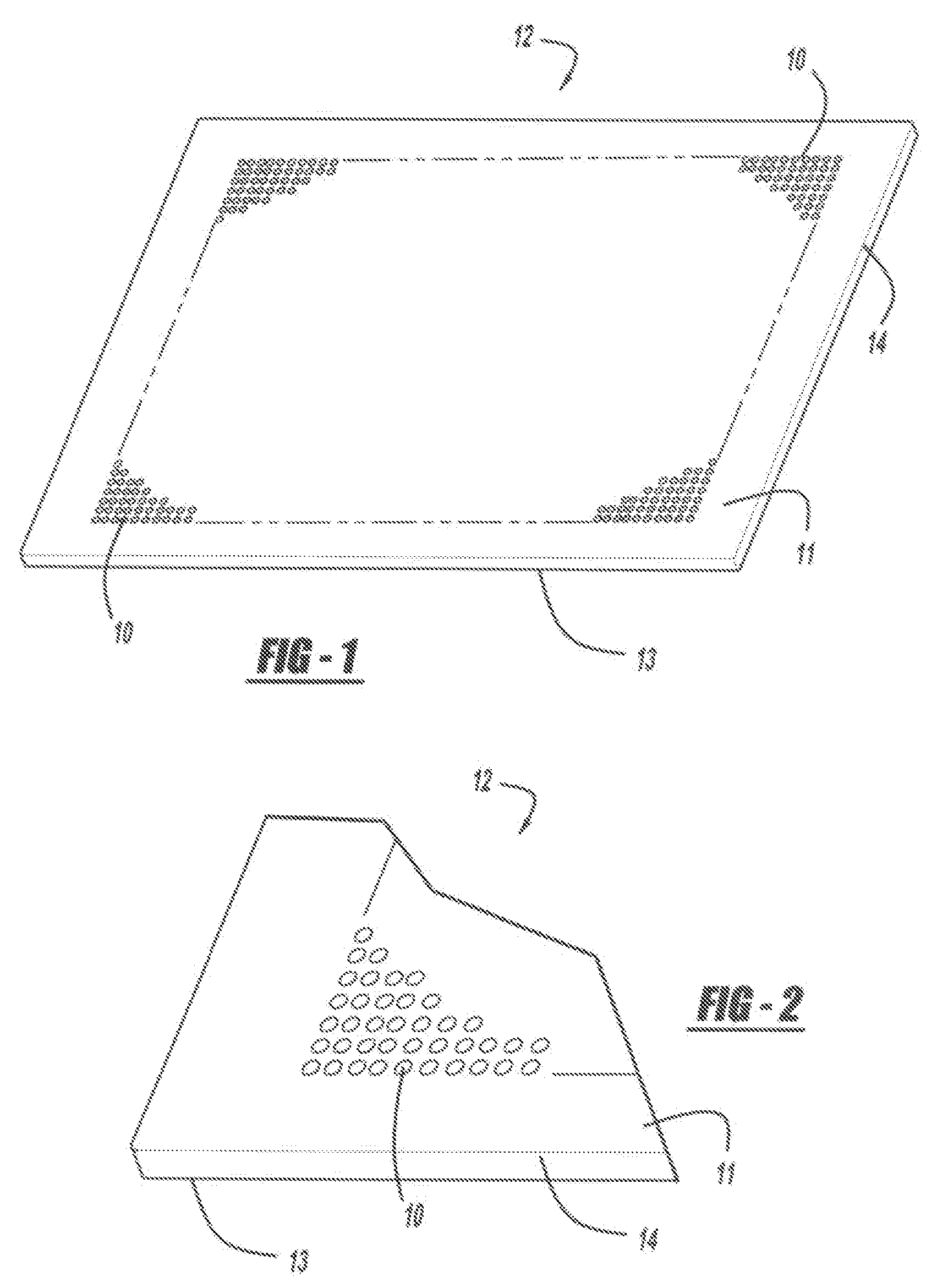
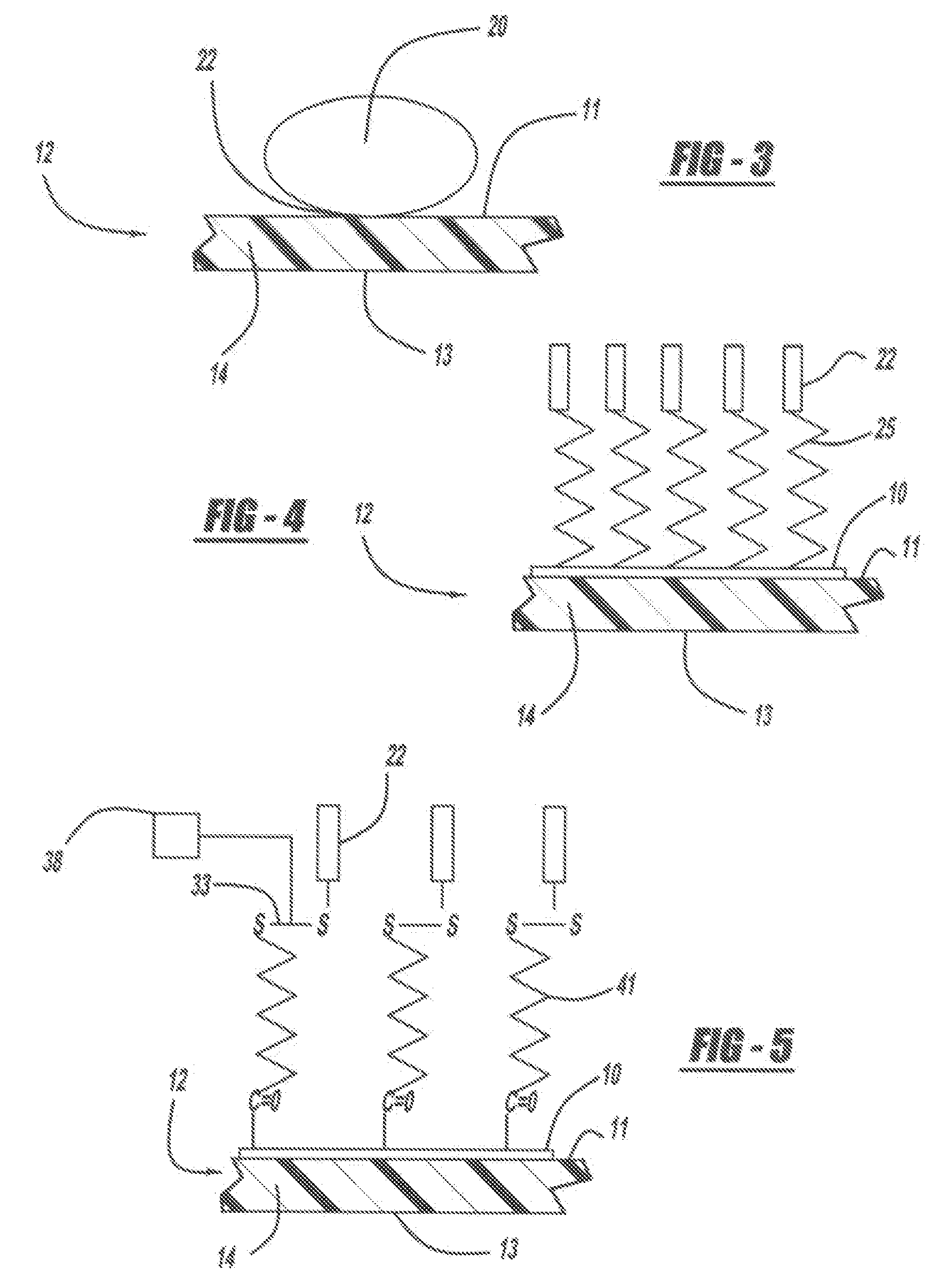
Optical disk-based assay devices and methods are described, in which analyte-specific signal elements are disposed on an optical disk substrate. In preferred embodiments, the analyte-specific signal elements are disposed readably with the disk's tracking features. Also described are cleavable signal elements particularly suitable for use in the assay device and methods. Binding of the chosen analyte simultaneously to a first and a second analyte-specific side member of the cleavable signal element tethers the signal-responsive moiety to the signal element's substrate-attaching end, despite subsequent cleavage at the cleavage site that lies intermediate the first and second side members. The signal responsive moiety reflects, absorbs, or refracts incident laser light. Described are nucleic acid hybridization assays, nucleic acid sequencing, immunoassays, cell counting assays, and chemical detection. Adaptation of the assay device substrate to function as an optical waveguide permits assay geometries suitable for continuous monitoring applications.
Owner:VINDUR TECH
Multiplexed measurement of membrane protein populations
Families of compositions are provided as labels, referred to as eTag reporters for attaching to polymeric compounds and assaying based on release of the eTag reporters from the polymeric compound and separation and detection. For oligonucleotides, the eTag reporters are synthesized at the end of the oligonucleotide by using phosphite or phosphate chemistry, whereby mass-modifying regions, charge-modifying regions and detectable regions are added sequentially to produce the eTag labeled reporters. By using small building blocks and varying their combination large numbers of different eTag reporters can be readily produced attached to a binding compound specific for the target compound of interest for identification. Protocols are used that release the eTag reporter when the target compound is present in the sample.
Owner:MONOGRAM BIOSCIENCES
Linkers and co-coupling agents for optimization of oligonucleotide synthesis and purification on solid supports
InactiveUS7211654B2Promote hydrolysisBioreactor/fermenter combinationsBiological substance pretreatmentsSolid surfaceTime-Consuming
A method of modulation of synthesis capacity on and cleavage properties of synthetic oligomers from solid support is described. The method utilizes linker molecules attached to a solid surface and co-coupling agents that have similar reactivities to the coupling compounds with the surface functional groups. The preferred linker molecules provide an increased density of polymers and more resistance to cleavage from the support surface. The method is particularly useful for synthesis of oligonucleotides, oligonucleotides microarrays, peptides, and peptide microarrays. The stable linkers are also coupled to anchor molecules for synthesis of DNA oligonucleotides using on support purification, eliminating time-consuming chromatography and metal cation presence. Oligonucleotides thus obtained can be directly used for mass analysis, DNA amplification and ligation, hybridization, and many other applications.
Owner:UNIV HOUSTON SYST
Compositions for sorting polynucleotides
InactiveUSRE39793E1Reduced stabilityAvoid precision problemsSequential/parallel process reactionsSugar derivativesPolynucleotideBiology
The invention provides a method of tracking, identifying, and / or sorting classes or subpopulations of molecules by the use of oligonucleotide tags. Oligonucleotide tags of the invention each consist of a plurality of subunits 3 to 6 nucleotides in length selected from a minimally cross-hybridizing set. A subunit of a minimally cross-hybridizing set forms a duplex or triplex having two or more mismatches with the complement of any other subunit of the same set. The number of oligonucleotide tags available in a particular embodiment depends on the number of subunits per tag and on the length of the subunit. An important aspect of the invention is the use of the oligonucleotide tags for sorting polynucleotides by specifically hybridizing tags attached to the polynucleotides to their complements on solid phase supports. This embodiment provides a readily automated system for manipulating and sorting polynucleotides, particularly useful in large-scale parallel operations, such as large-scale DNA sequencing, mRNA fingerprinting, and the like, wherein many target polynucleotides or many segments of a single target polynucleotide are sequenced simultaneously.
Owner:SOLEXA
Peptide display arrays
InactiveUS20120270748A1Improve throughputDone relatively cheaplyNucleotide librariesAntibody mimetics/scaffoldsPeptide displayDiscrete set
The present invention provides arrays, methods of constructing arrays, and methods of use of such arrays. The arrays of the invention comprise a substrate with two or more discrete constructs or discrete sets of constructs associated on the surface of the substrate, optionally via a linker molecule. The constructs include an oligonucleotide comprising a region encoding a peptide of interest and an affinity tag and an untranslated region, a fusion peptide comprising both the peptide of interest and the affinity tag and a capture agent that forms a binding pair with the affinity tag.
Owner:PROGNOSYS BIOSCI
Pre-anchor wash
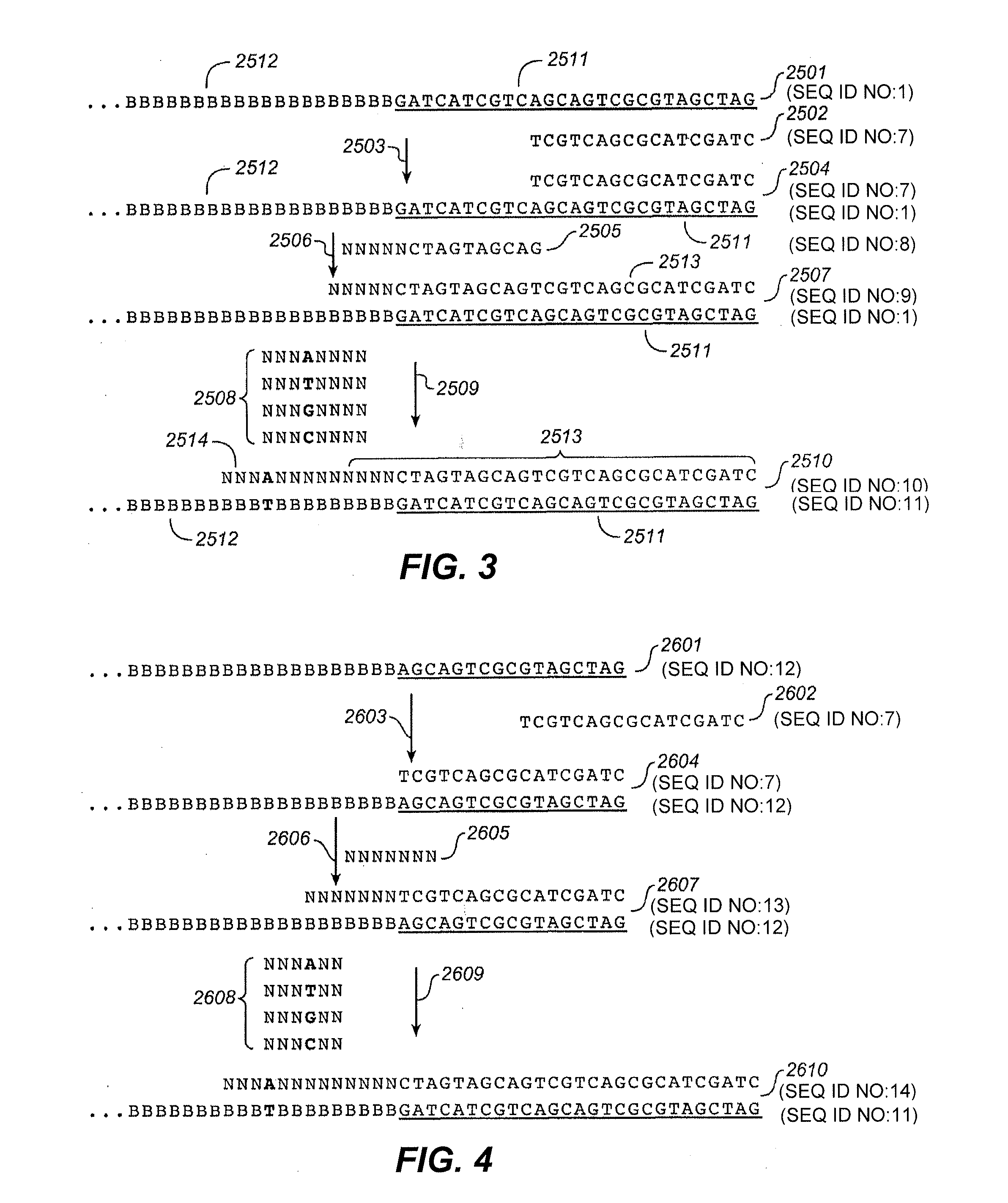
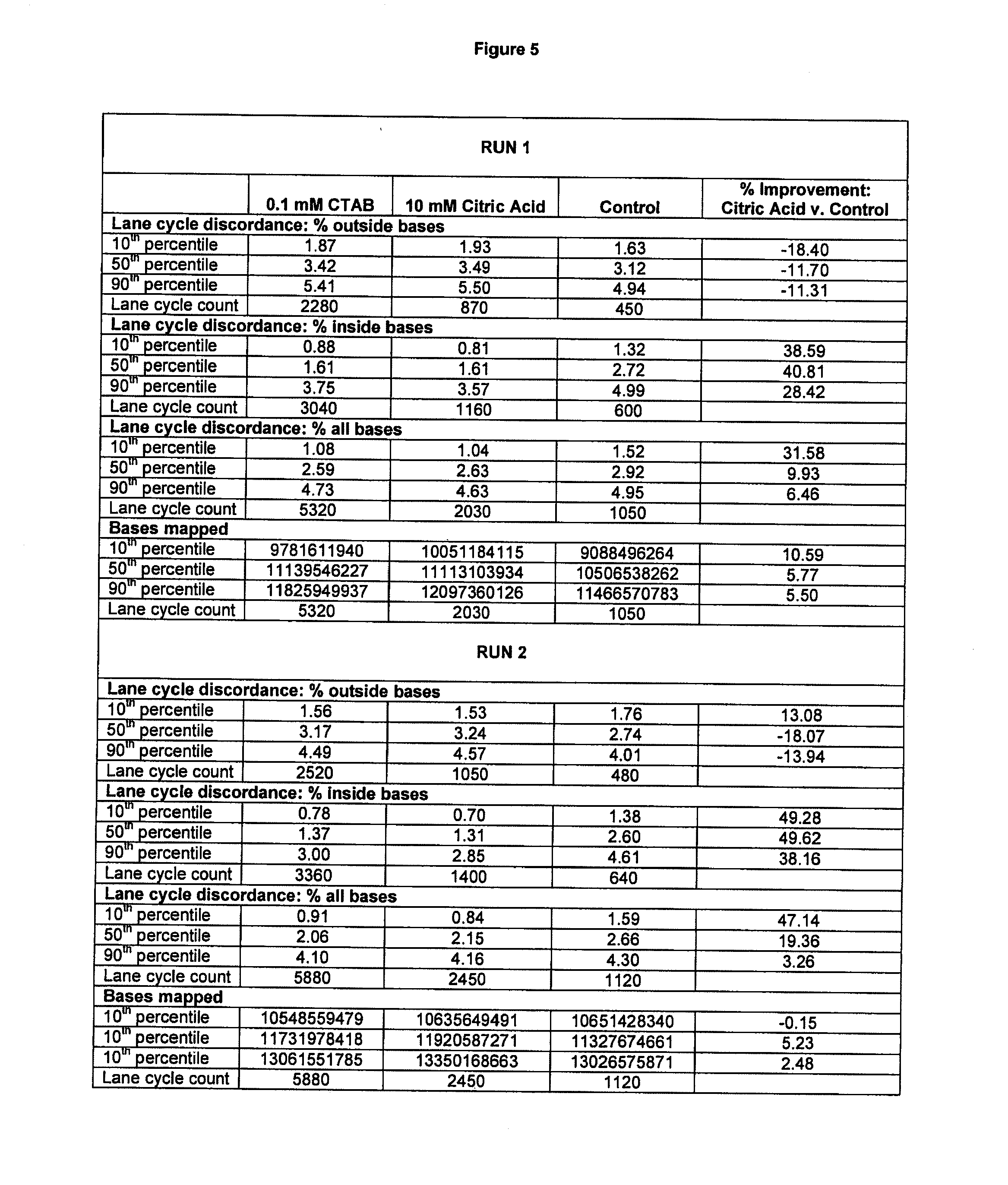
InactiveUS20130296173A1Reduce inconsistencyIncrease mappable yieldMicrobiological testing/measurementLibrary linkersNucleic acid sequencingComputational biology
The present invention is directed to compositions and methods for improving the discordance rate and mapping yield in nucleic acid sequencing reactions.
Owner:COMPLETE GENOMICS INC
Methods and compositions for making locus-specific arrays
InactiveUS20050136414A1Bioreactor/fermenter combinationsBiological substance pretreatmentsCell biologySugar nucleotide
Owner:ILLUMINA INC
Micro-circuit system with array of functionalized micro-electrodes
InactiveUS6203758B1Immobilised enzymesSequential/parallel process reactionsCombinatorial chemistryElectron
A micro-circuit for performing analyses of multimolecular interactions and for performing molecular syntheses, comprising: (a) a support; (b) at least one micro-electrode attached to the support, the micro-electrode being selectively electronically activated and the micro-electrode having a protective layer which is removable; (c) a binding entity for attachment to the at least one micro-electrode, the binding entity being capable of attachment to at least one micro-electrode when the protective layer has been removed; and (d) a power source being operatively connected to at least one micro-electrode for electronically activating at least one micro-electrode. The micro-circuit of the present invention also includes embodiments featuring a micro-circuit reader for detecting the interaction of the binding entity to a complementary probe, as well as methods for making and using the micro-circuit of the present invention.
Owner:BEN GURION UNIVERSITY OF THE NEGEV
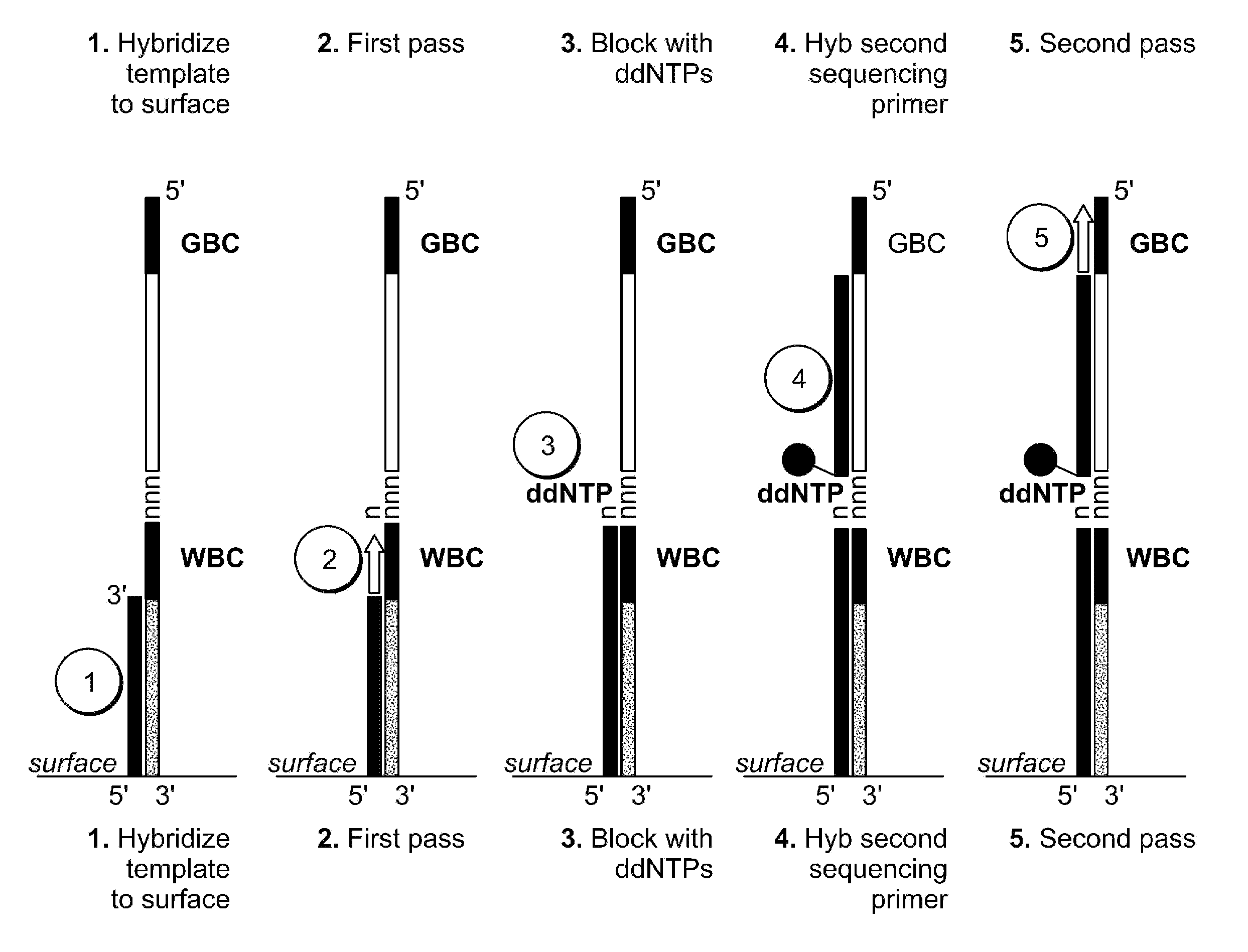
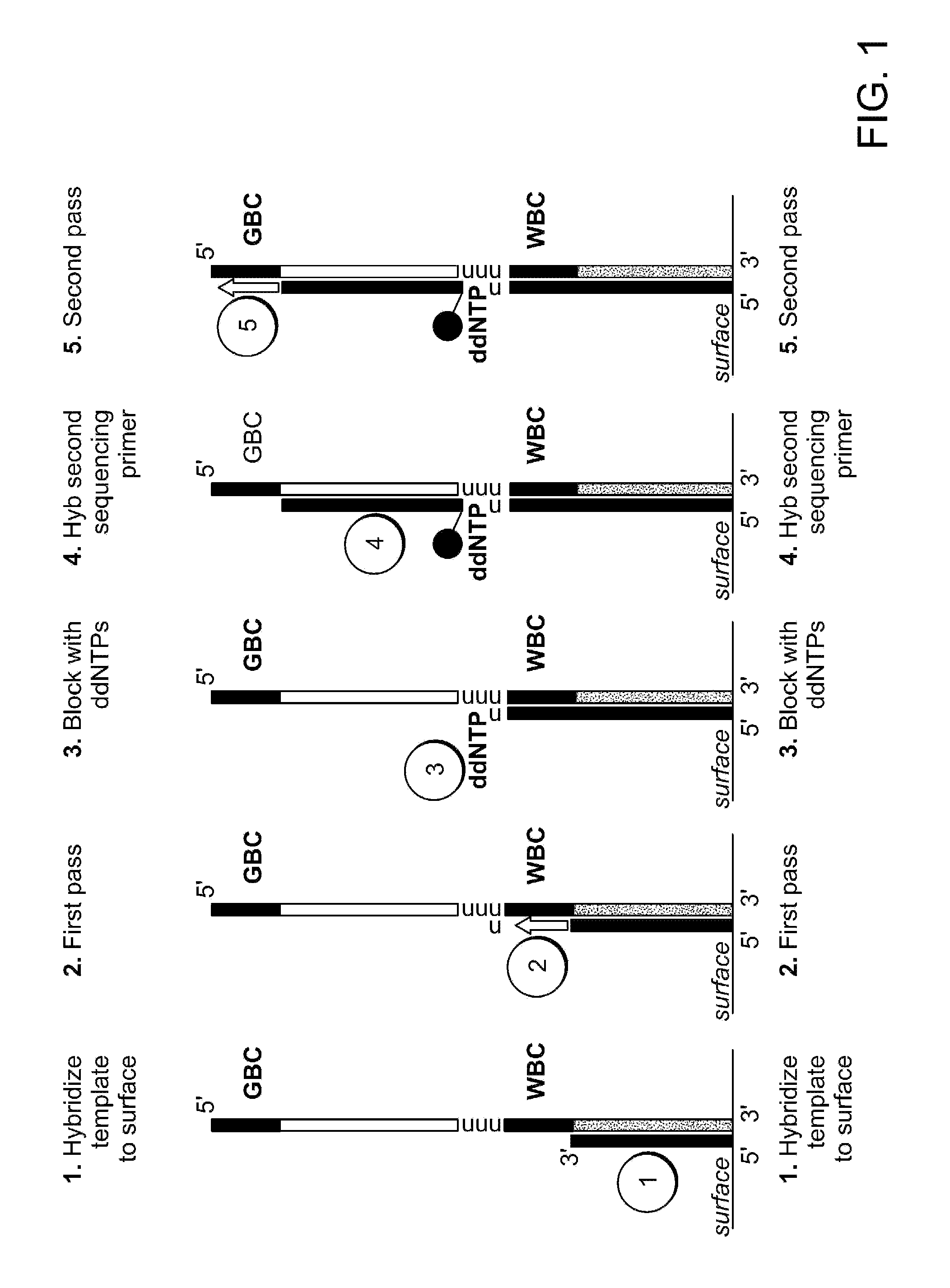
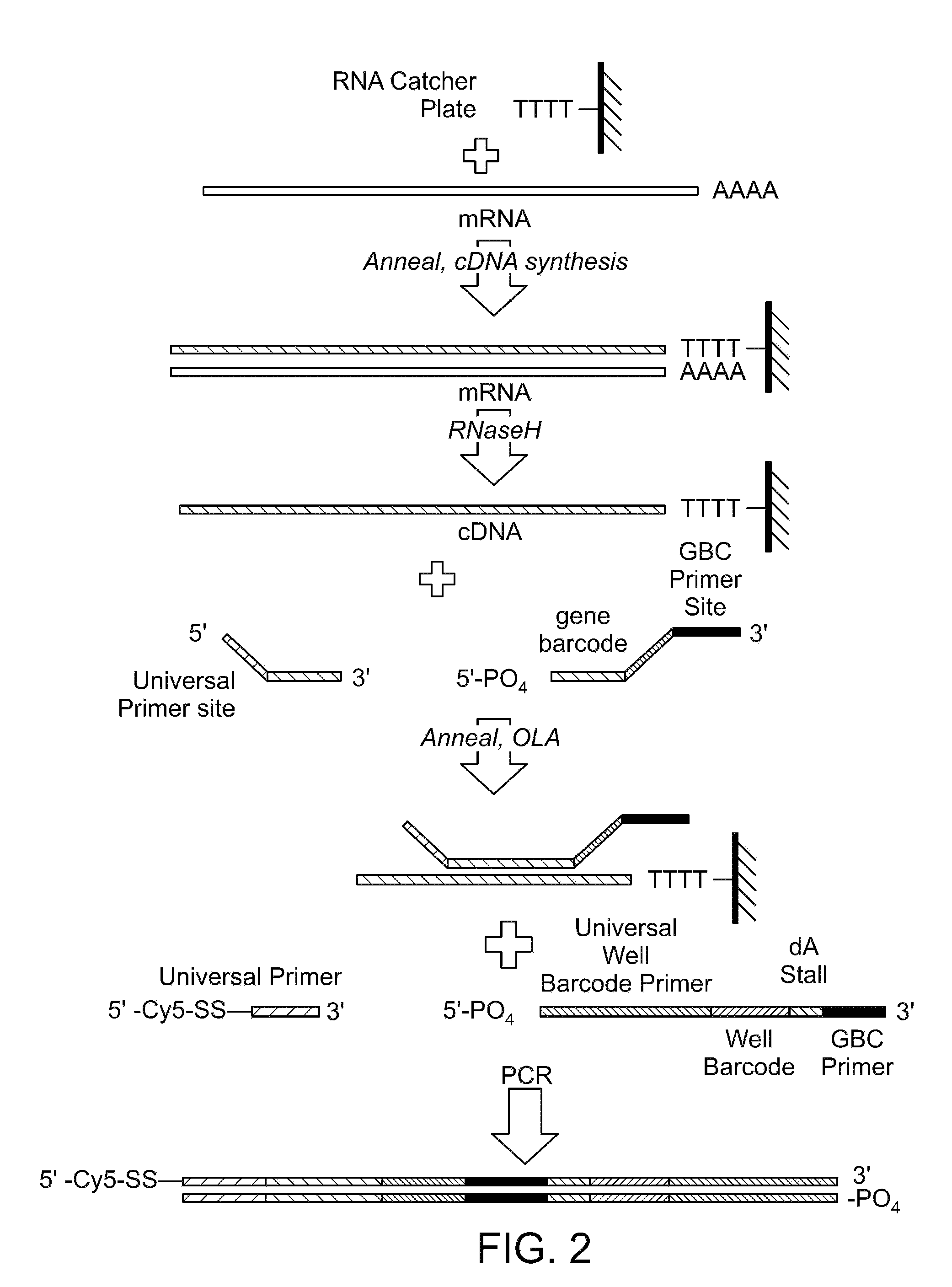
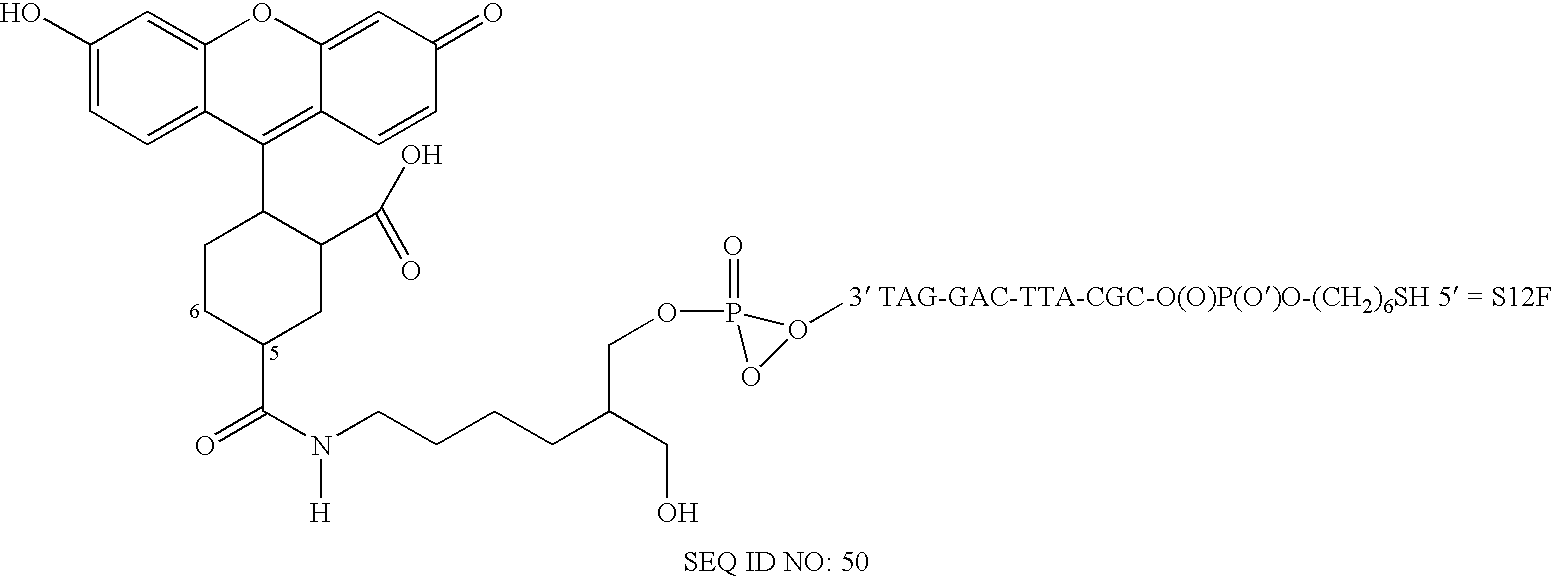
Two-primer sequencing for high-throughput expression analysis
InactiveUS20090163366A1Improve efficiencyImprove accuracySugar derivativesLibrary tagsBarcodePolynucleotide
Owner:FLUIDIGM CORP
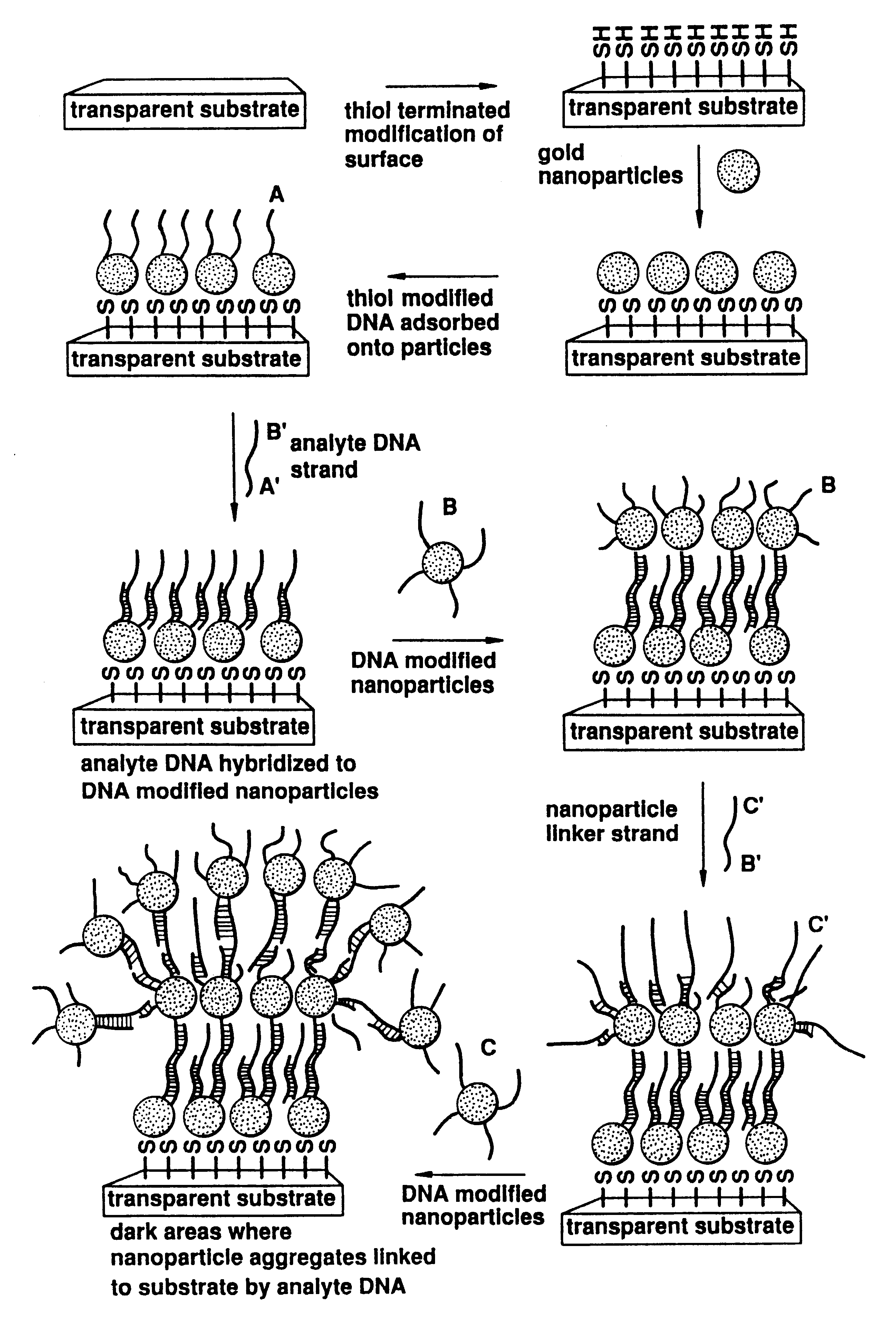
Nanoparticles having oligonucleotides attached thereto and uses therefor
InactiveUS6984491B2Easy to moveImprove bindingMaterial nanotechnologySugar derivativesNanoparticleColor changes
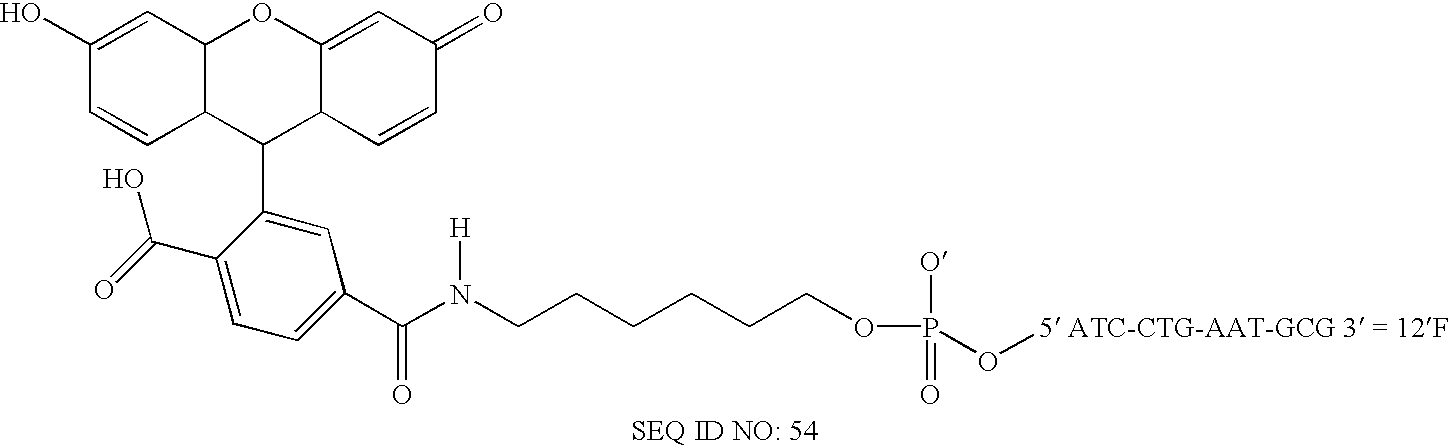
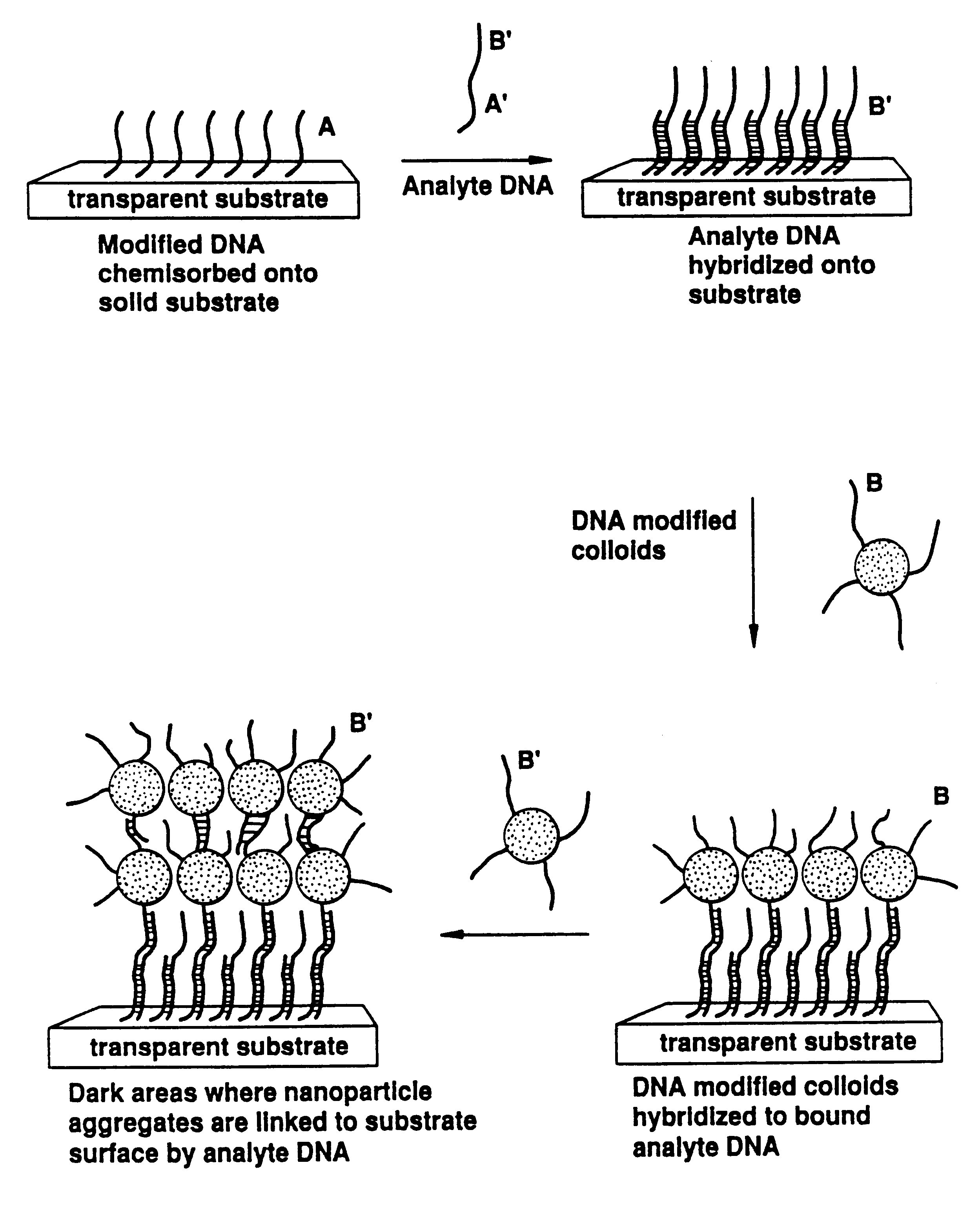
The invention provides methods of detecting a nucleic acid. The methods comprise contacting the nucleic acid with one or more types of particles having oligonucleotides attached thereto. In one embodiment of the method, the oligonucleotides are attached to nanoparticles and have sequences complementary to portions of the sequence of the nucleic acid. A detectable change (preferably a color change) is brought about as a result of the hybridization of the oligonucleotides on the nanoparticles to the nucleic acid. The invention also provides compositions and kits comprising particles. The invention further provides methods of synthesizing unique nanoparticle-oligonucleotide conjugates, the conjugates produced by the methods, and methods of using the conjugates. In addition, the invention provides nanomaterials and nanostructures comprising nanoparticles and methods of nanofabrication utilizing nanoparticles. Finally, the invention provides a method of separating a selected nucleic acid from other nucleic acids.
Owner:NORTHWESTERN UNIV +1
Methods for synthesis of encoded libraries
The present invention provides a method of synthesizing libraries of molecules which include an encoding oligonucleotide tag.
Owner:GLAXO SMITHKLINE LLC
Optical disk-based assay devices and methods
InactiveUS20020106661A1Avoid lostStrong specificitySequential/parallel process reactionsMaterial analysis by optical meansAnalyteLaser light
Optical disk-based assay devices and methods are described, in which analyte-specific signal elements are disposed on an optical disk substrate. In preferred embodiments, the analyte-specific signal elements are disposed readably with the disk's tracking features. Also described are cleavable signal elements particularly suitable for use in the assay device and methods. Binding of the chosen analyte simultaneously to a first and a second analyte-specific side member of the cleavable signal element tethers the signal-responsive moiety to the signal element's substrate-attaching end, despite subsequent cleavage at the cleavage site that lies intermediate the first and second side members. The signal responsive moiety reflects, absorbs, or refracts incident laser light. Described are nucleic acid hybridization assays, nucleic acid sequencing, immunoassays, cell counting assays, and chemical detection. Adaptation of the assay device substrate to function as an optical waveguide permits assay geometries suitable for continuous monitoring applications.
Owner:NAGAOKA
Combinatorial Electrochemical Synthesis
<heading lvl="0">Abstract of Disclosure< / heading> An array of selectively addressible microelectrodes for combinatorial synthesis of complex polymers or alloys.
Owner:THERASENSE
Functionalized polydiacetylene sensors
ActiveUS20110059867A1Improve conductivityPeptide librariesPeptide/protein ingredientsAnalyteCoupling
A microarray includes a solid substrate having a surface, the surface having a plurality of binding spots and a plurality of reaction moieties bound to the binding spots. A reaction moiety includes a plurality of polyacetylene monomers, the polyacetylene monomers having a first coupling region and a second coupling region, the first coupling region having a first functional group operable to bind to the binding spot and the second coupling region comprising a second functional group operable to bind to an accessory molecule; and an accessory molecule having a binding region and an analyte reaction region, the analyte reaction region operable to selectively bind to the target analyte, and the binding region operable to bind to the second coupling region of the polyacetylene monomer. Upon binding a target analyte with the reaction moiety, a color change from the polyacetylene monomer occurs and the reaction moiety produces fluorescence.
Owner:RGT UNIV OF MICHIGAN
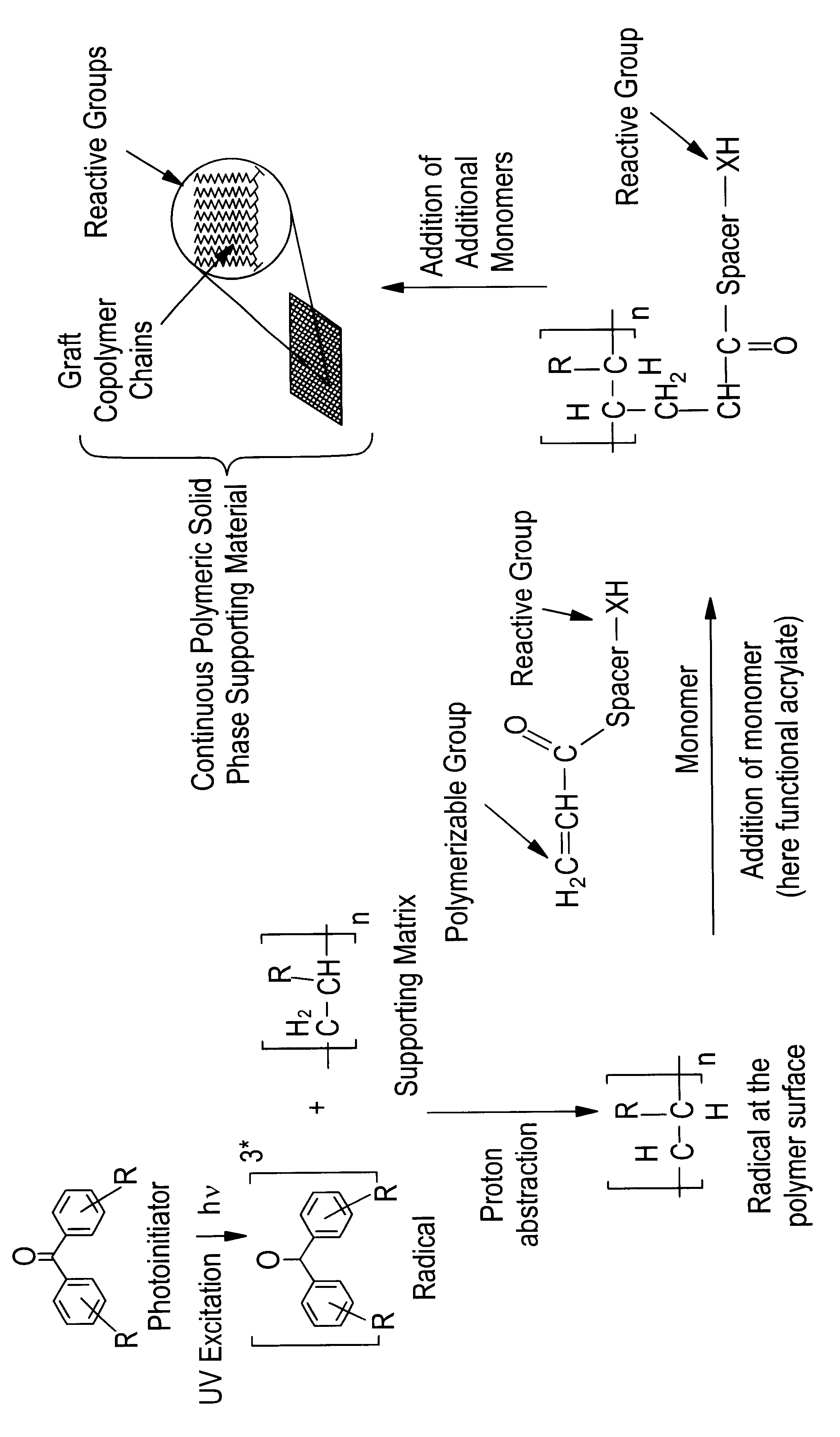
Method for the parallel and combinatory synthesis of compounds bound to a continuous polymeric solid phase supporting material
InactiveUS6515039B1Improve performanceImprove accessibilitySequential/parallel process reactionsMacromolecular librariesSimple Organic CompoundsPolymer science
The invention relates to a method for the parallel and combinatory synthesis of compounds bound to a continuous polymeric solid phase supporting material. According to the method, a continuous polymeric solid phase supporting material is prepared, the material comprising a matrix of a supporting polymeric material and graft copolymer chains covalently bound to the supporting matrix, the graft copolymer chains having reactive groups being able to react with organic compounds, thereby forming spatially defined reaction sites. The method then comprises the sequential spotting of synthesis building blocks or reagents at the various reaction sites on the continuous solid phase supporting material to yield a substrate library of compounds bound to the solid phase supporting material, whereby each reaction site on the continuous solid phase supporting material determines the composition of the synthesized compound, which is bound directly on the supporting matrix via the reactive groups of the graft polymer chain.
Owner:POLY AN
Preparation and use of mixed mode solid substrates for chromatography adsorbents and biochip arrays
InactiveUS7144743B2Improve adsorption capacityIon-exchanger regenerationLibrary member identificationPhosphateSorbent
For certain mixed mode resins having anionic character, a ligand is joined to a solid support via a linkage that includes a mercapto-, ether- or amino-containing moiety. A suitable ligand comprises an aromatic group, a heteroaromatic group, or a heterocyclic group, optionally fused, that is sulfate-, sulfonate-, phosphonate- or phosphate-substituted and that is linked to such a moiety. These resins possess an anionic character under conditions prescribed for their use. Separation of a biological substance, such as a peptide or protein, can be accomplished with a resin of this type via a change in the pH of eluants, thereby effecting adsorption and desorption.
Owner:PALL CORP
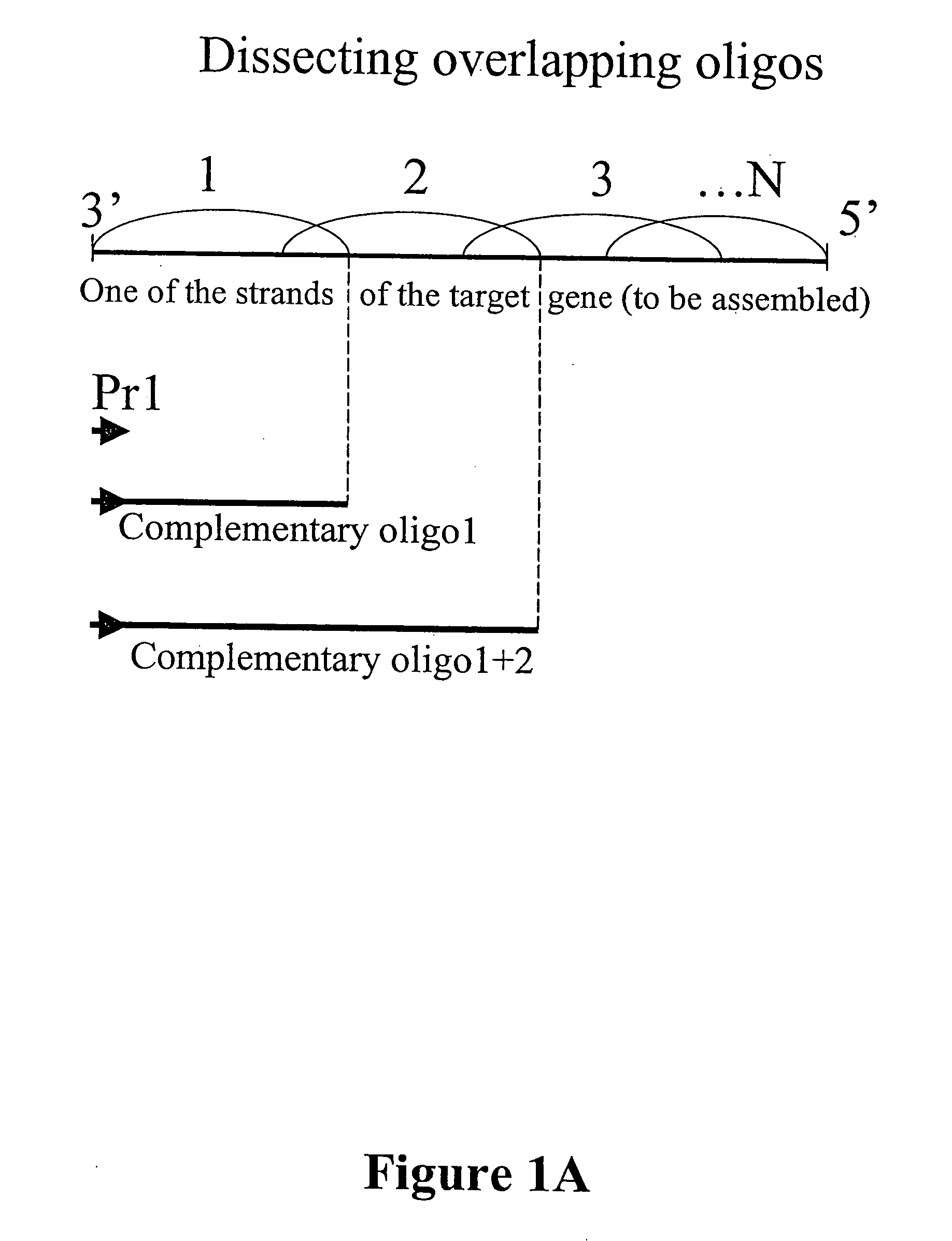
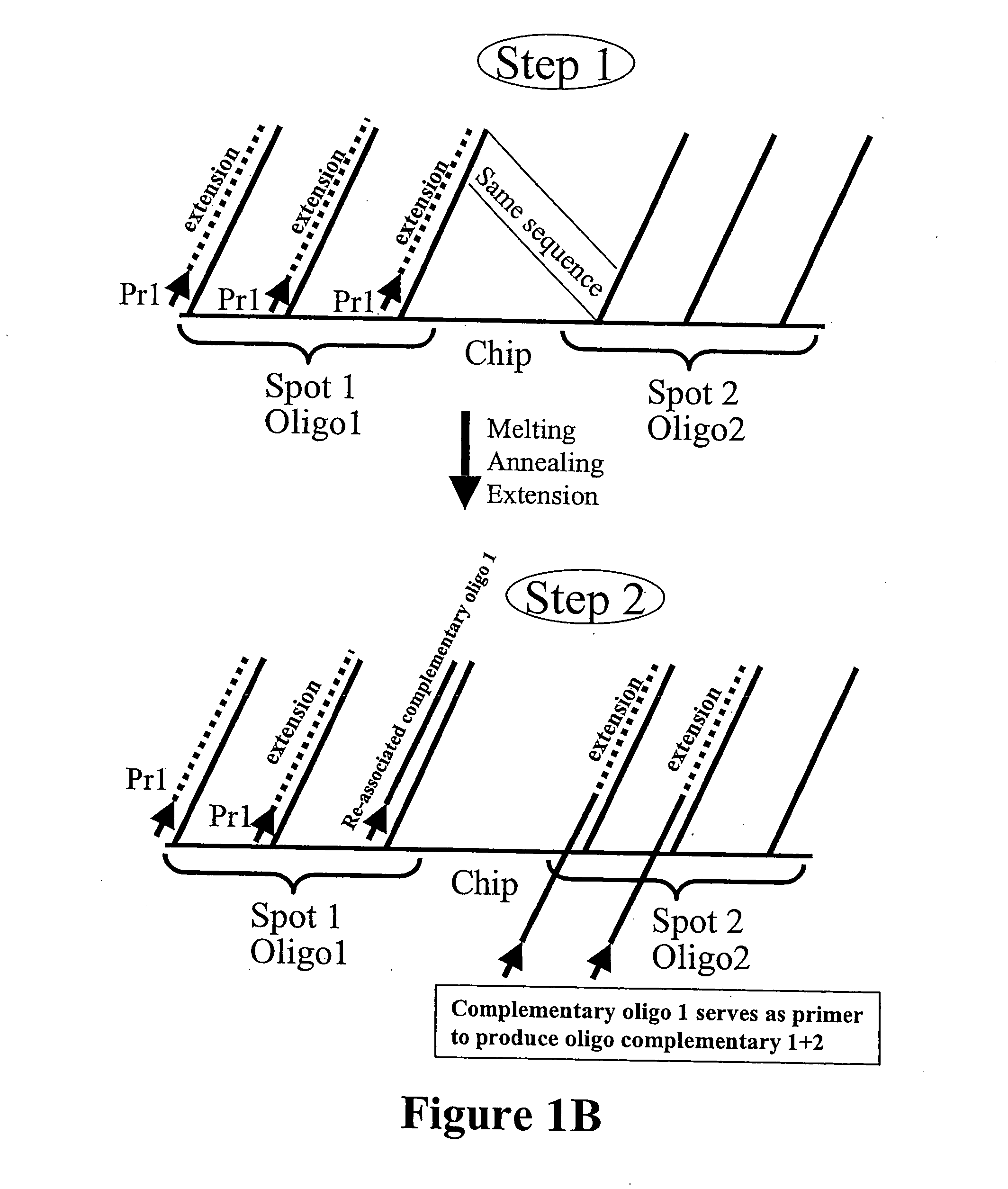
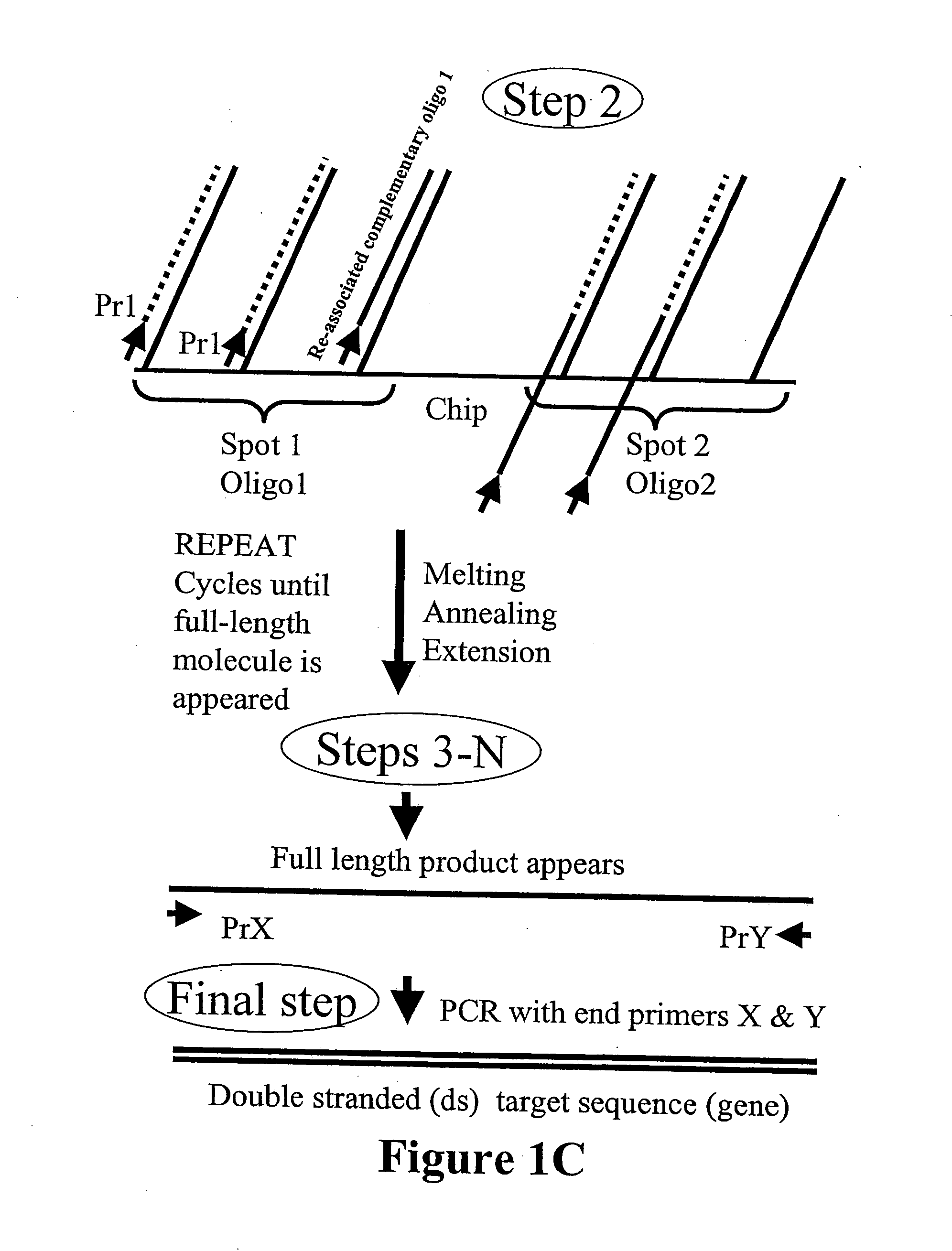
Microarray synthesis and assembly of gene-length polynucleotides
InactiveUS20050287585A1Material nanotechnologySequential/parallel process reactionsPolynucleotideOligonucleotide
There is disclosed a process for in vitro synthesis and assembly of long, gene-length polynucleotides based upon assembly of multiple shorter oligonucleotides synthesized in situ on a microarray platform. Specifically, there is disclosed a process for in situ synthesis of oligonucleotide fragments on a solid phase microarray platform and subsequent, “on device” assembly of larger polynucleotides composed of a plurality of shorter oligonucleotide fragments.
Owner:GEN9
Preparation and screening of crystalline zeolite and hydrothermally-synthesized materials
Methods and apparatus for the preparation and use of a substrate having an array of diverse materials in predefined regions thereon. A substrate having an array of diverse materials thereon is generally prepared by delivering components of materials to predefined regions on a substrate, and simultaneously reacting the components to form at least two materials. Materials which can be prepared using the methods and apparatus of the present invention include, for example, covalent network solids, ionic solids and molecular solids. More particularly, materials which can be prepared using the methods and apparatus of the present invention include, for example, inorganic materials, intermetallic materials, metal alloys, ceramic materials, organic materials, organometallic materials, non-biological organic polymers, composite materials (e.g., inorganic composites, organic composites, or combinations thereof), etc. Once prepared, these materials can be screened for useful properties including, for example, electrical, thermal, mechanical, morphological, optical, magnetic, chemical, or other properties. Thus, the present invention provides methods for the parallel synthesis and analysis of novel materials having useful properties.
Owner:INTERMOLECULAR
Microarray synthesis and assembly of gene-length polynucleotides
InactiveUS20060035218A1Material nanotechnologySequential/parallel process reactionsPolynucleotideGene
Owner:GEN9
Combinatorial synthesis and screening of non-biological polymers
Methods and apparatus for the preparation and use of a substrate having an array of diverse materials in predefined regions thereon. A substrate having an array of diverse materials thereon is generally prepared by delivering components of materials to predefined regions on a substrate, and simultaneously reacting the components to form at least two materials. Materials which can be prepared using the methods and apparatus of the present invention include, for example, covalent network solids, ionic solids and molecular solids. More particularly, materials which can be prepared using the methods and apparatus of the present invention include, for example, inorganic materials, intermetallic materials, metal alloys, ceramic materials, organic materials, organometallic materials, non-biological organic polymers, composite materials (e.g., inorganic composites, organic composites, or combinations thereof), etc. Once prepared, these materials can be screened for useful properties including, for example, electrical, thermal, mechanical, morphological, optical, magnetic, chemical, or other properties. Thus, the present invention provides methods for the parallel synthesis and analysis of novel materials having useful properties.
Owner:INTERMOLECULAR
Composite microarray slides
InactiveUS20030219816A1Useful characteristicBioreactor/fermenter combinationsPeptide librariesMicroscope slideBiopolymer
Improved composite microarray slides for use in micro-analytical diagnostic applications are disclosed. Specifically, composite microarray slides useful for carrying a microarray of biological polymers on the surface thereof including composite microarray slides having a porous membrane formed by a phase inversion process effectively attached by covalent bonding through chemical agents that comprise anchor / linker moieties to a substrate that prepares the substrate to sufficiently bond to the porous membrane formed by a phase inversion process such that the combination produced thereby is useful in microarray applications and wherein the composite microarray slides are covalently bonded to a solid base member, such as, for example, a glass or Mylar microscope slide, such that the combination produced thereby is useful in microarray applications. Apparatus and methods for fabricating the composite microarray slides are also disclosed.
Owner:3M INNOVATIVE PROPERTIES CO
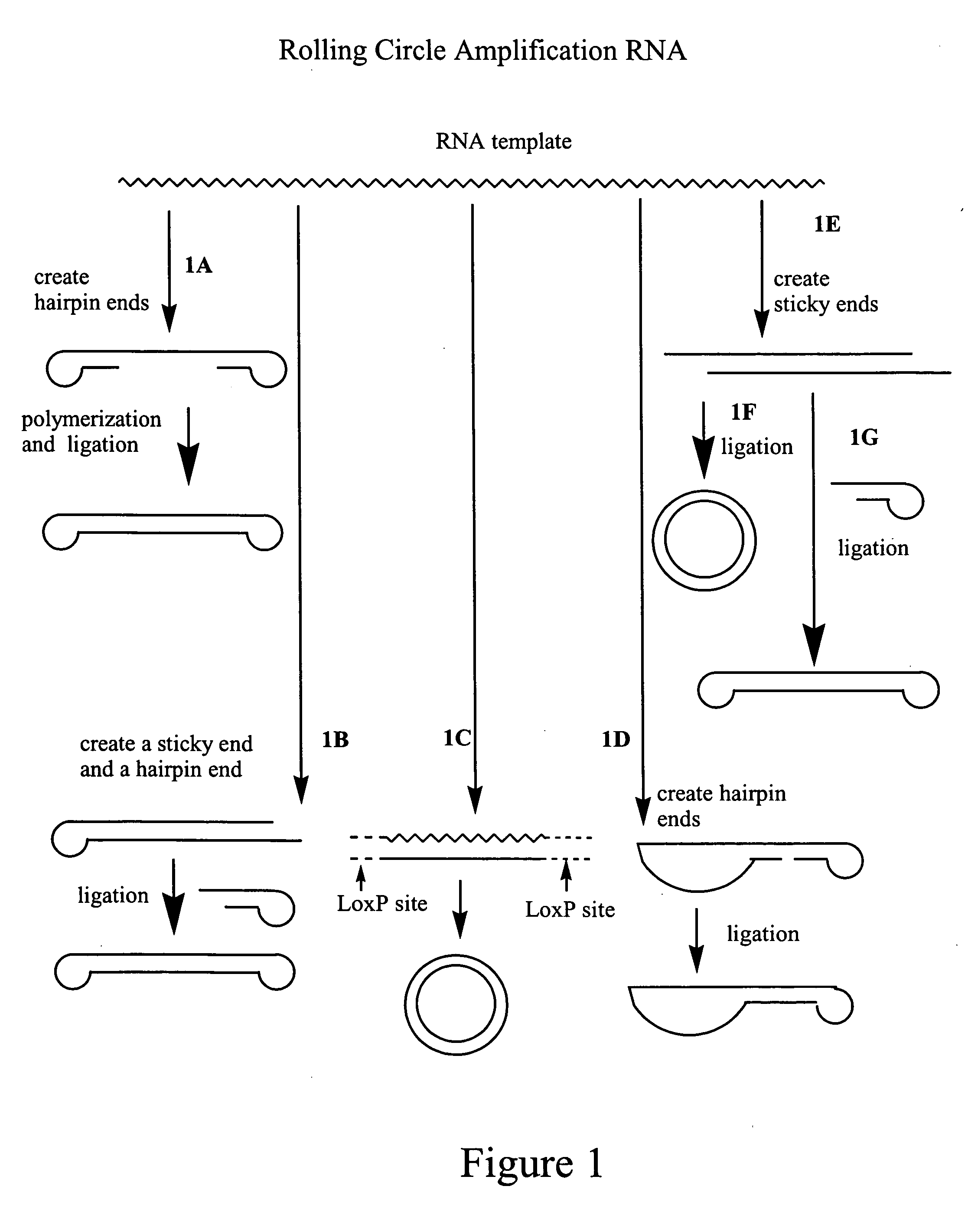
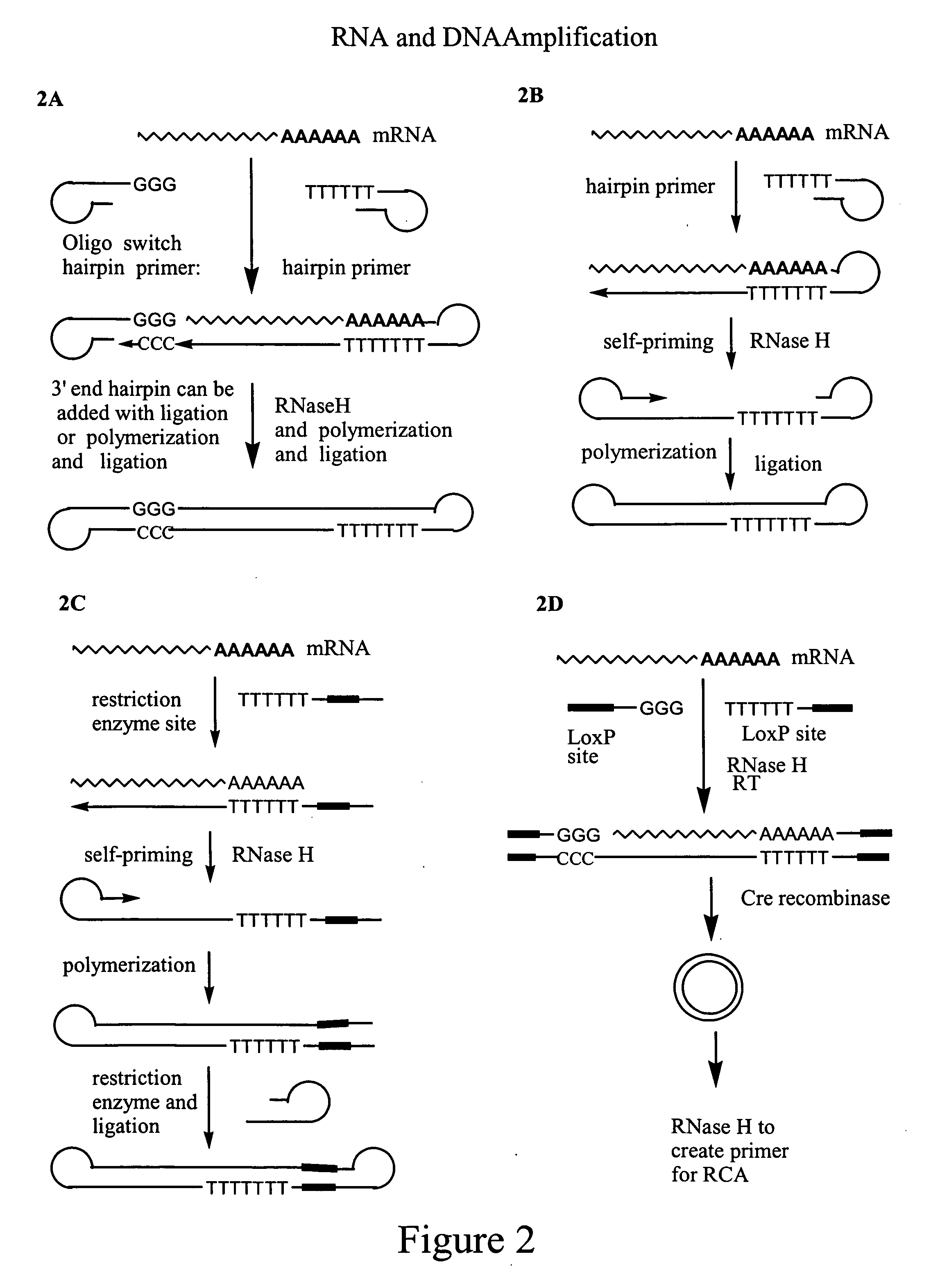
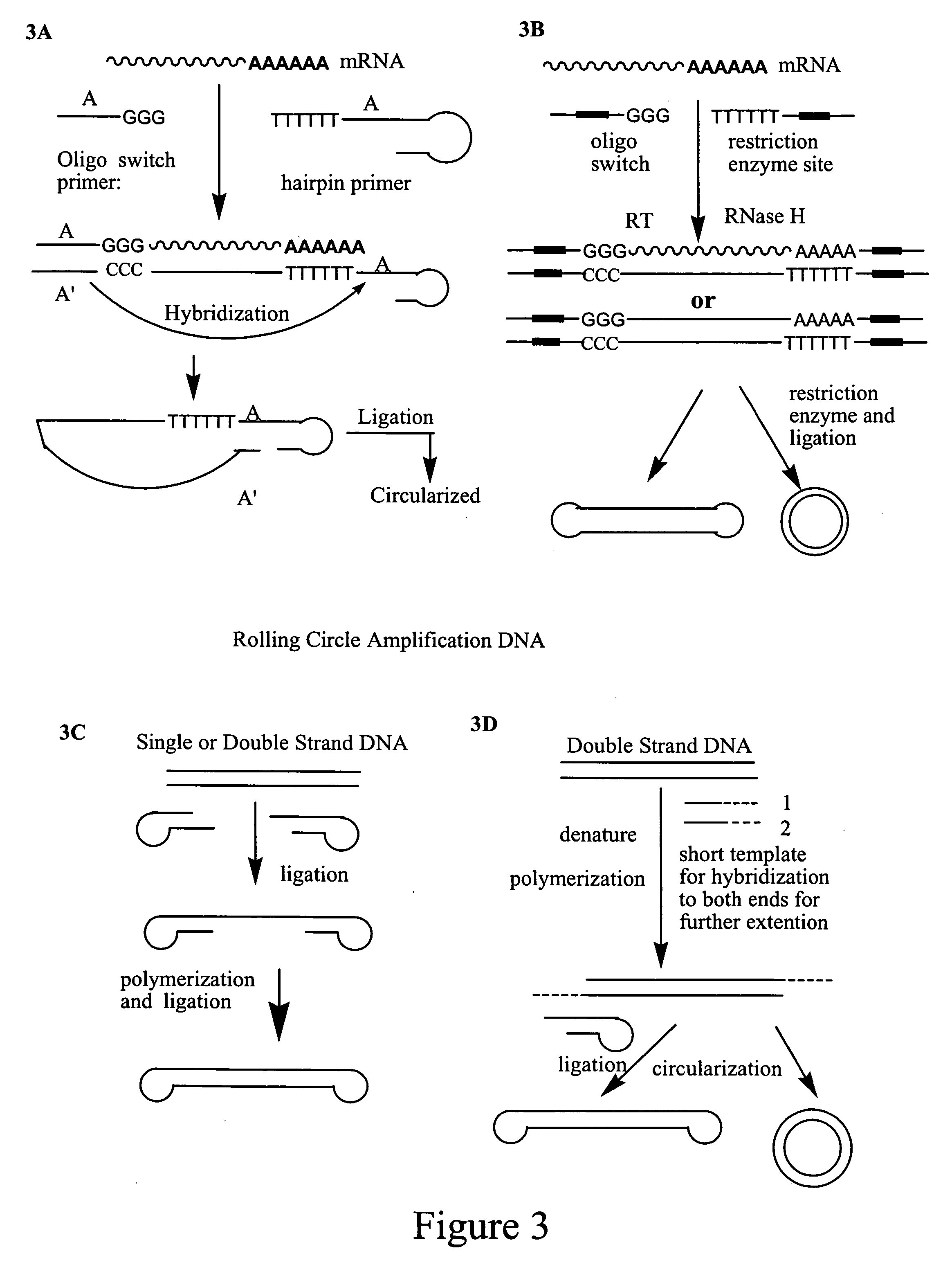
Amplification of polynucleotides by rolling circle amplification
InactiveUS20050069938A1Improve responseMicrobiological testing/measurementLibrary linkersNucleic acid sequencingPolynucleotide
Rolling circle amplification is used to amplify and detect target nucleic acid molecules by affixing a first and / or second linker nucleic acid molecule or a second linker nucleic acid molecule to the target nucleic acid molecule, then circularizing the target nucleic acid molecule, and then amplifying the circularized nucleic acid molecule to generate reiterated nucleic acid sequences. The methods may be used to amplify nucleic acids, particularly RNA, for detection and cloning.
Owner:CIRCLEAMP
Nanoparticles having oligonucleotides attached thereto and uses therefor
The invention provides methods of detecting a nucleic acid. The methods comprise contacting the nucleic acid with one or more types of particles having oligonucleotides attached thereto. In one embodiment of the method, the oligonucleotides are attached to nanoparticles and have sequences complementary to portions of the sequence of the nucleic acid. A detectable change (preferably a color change) is brought about as a result of the hybridization of the oligonucleotides on the nanoparticles to the nucleic acid. The invention also provides compositions and kits comprising particles. The invention further provides methods of synthesizing unique nanoparticle-oligonucleotide conjugates, the conjugates produced by the methods, and methods of using the conjugates. In addition, the invention provides nanomaterials and nanostructures comprising nanoparticles and methods of nanofabrication utilizing nanoparticles. Finally, the invention provides a method of separating a selected nucleic acid from other nucleic acids.
Owner:NORTHWESTERN UNIV +1
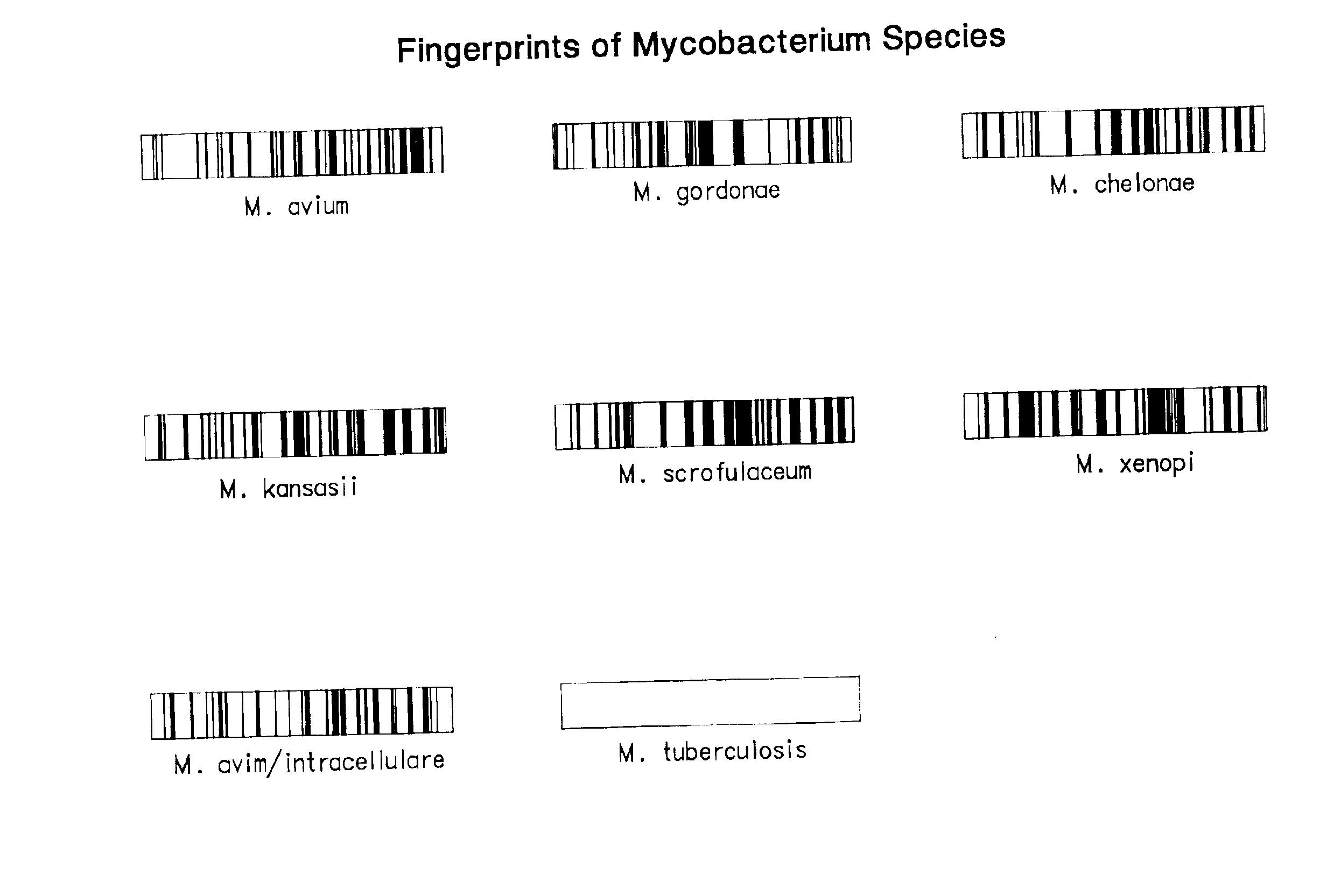
Chip-based species identification and phenotypic characterization of microorganisms
This invention provides oligonucleotide based arrays and methods for speciating and phenotyping organisms, for example, using oligonucleotide sequences based on the Mycobacterium tubercluosis rpoB gene. The groups or species to which an organism belongs may be determined by comparing hybridization patterns of target nucleic acid from the organism to hybridization patterns in a database.
Owner:AFFYMETRIX INC
Protein arrays
InactiveUS7642085B2Easy to detectEasy to quantifyBioreactor/fermenter combinationsOrganic active ingredientsCombinatorial chemistryAmino acid
The invention provides proteins attached to solid supports, and methods of preparing such solid support-bound proteins are provided. The proteins are attached to solid supports by means of an unnatural amino acid incorporated into the protein, which unnatural amino acid includes a reactive group that can react with a second reactive group that is attached to a solid support.
Owner:THE SCRIPPS RES INST
Alpha-isocyanocarboxylate solid support templates, method of preparation and for using the same
InactiveUS20040127719A1Silicon organic compoundsOrganic compound preparationSolid reactionCombinatorial chemistry
A solid phase reaction template for the production of heterocyclic scaffolds in a reaction media, said solid phase component comprising an alpha-isocyanocarboxylate core compound linked to a solid substrate, the template having the formula: A method for the production of heterocyclic scaffolds using the template is disclosed
Owner:ADVANCED SYNTECH
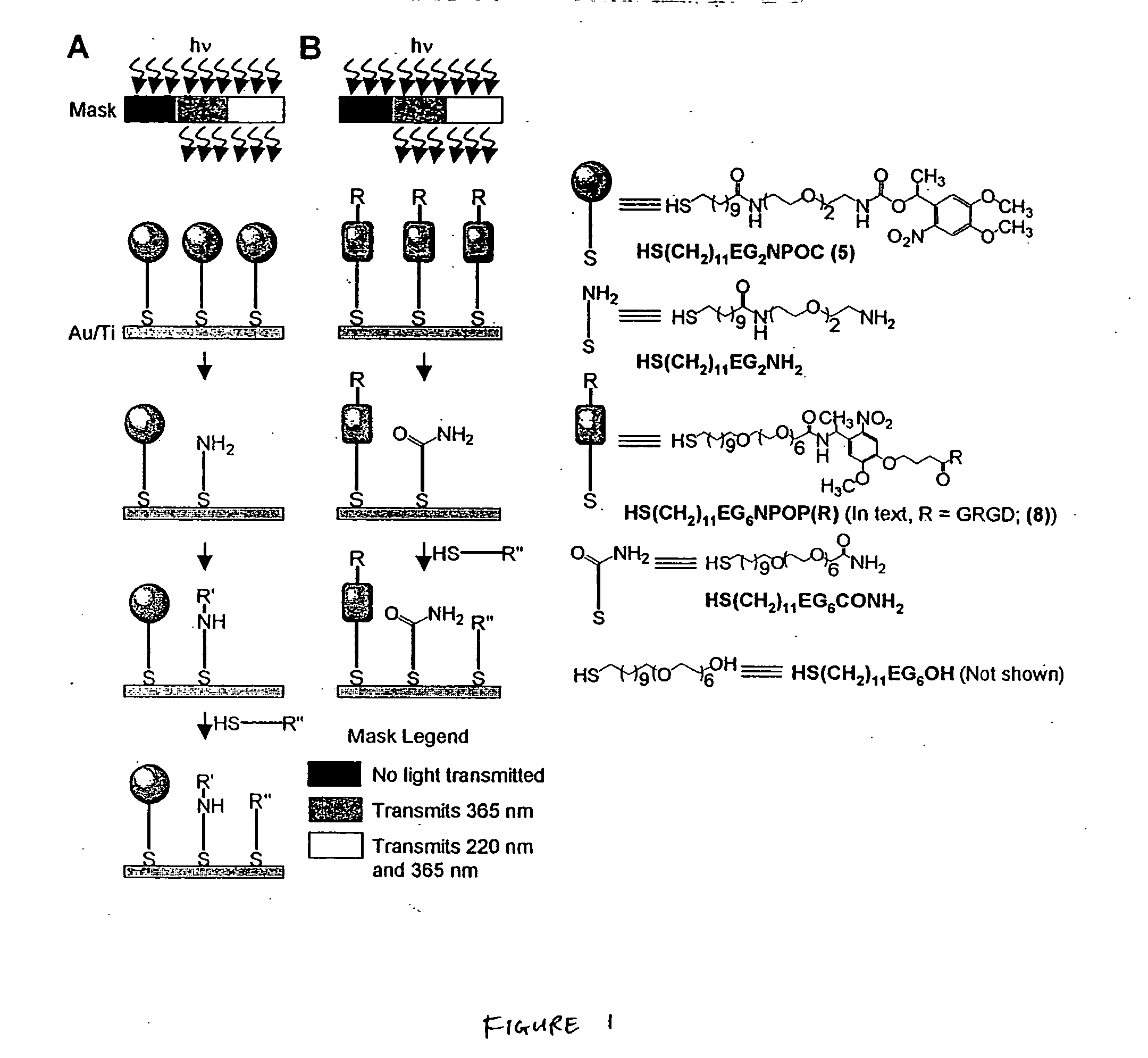
Patterning and alteration of molecules
The present invention provides a series of methods, compositions, and articles for patterning a surface with multiple, aligned layers of molecules, by exposing the molecules to electromagnetic radiation. In certain embodiments, a single photomask acts as an area-selective filter for light at multiple wavelengths. A single set of exposures of multiple wavelengths through this photomask may make it possible to fabricate a pattern comprising discontinuous multiple regions, where the regions differ from each other in at least one chemical and / or physical property, without acts of alignment between the exposures. In certain embodiments, the surface includes molecules attached thereto that can be photocleaved upon exposure to a certain wavelength of radiation, thereby altering the chemical composition on at least a portion of the surface. In some embodiments, the molecules attached to the surface may include thiol moieties (e.g., as in alkanethiol), by which the molecule can become attached to the surface. In some embodiments, the molecules may be terminated at the unattached end with photocleavable groups. In other embodiments, a molecule that was photocleaved may be exposed to another molecule that binds to the photocleaved molecule. In certain cases, the molecules may be terminated at the unattached end with hydrophilic groups that may, for example, be resistant to the adsorption of proteins. In other cases, the molecules may be terminated at the unattached end with end groups that are not resistant to the adsorption of proteins. In certain embodiments, the techniques are used to pattern simultaneously two different regions that are resistant to the adsorption of proteins, and a third region that does not resist the adsorption of proteins.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
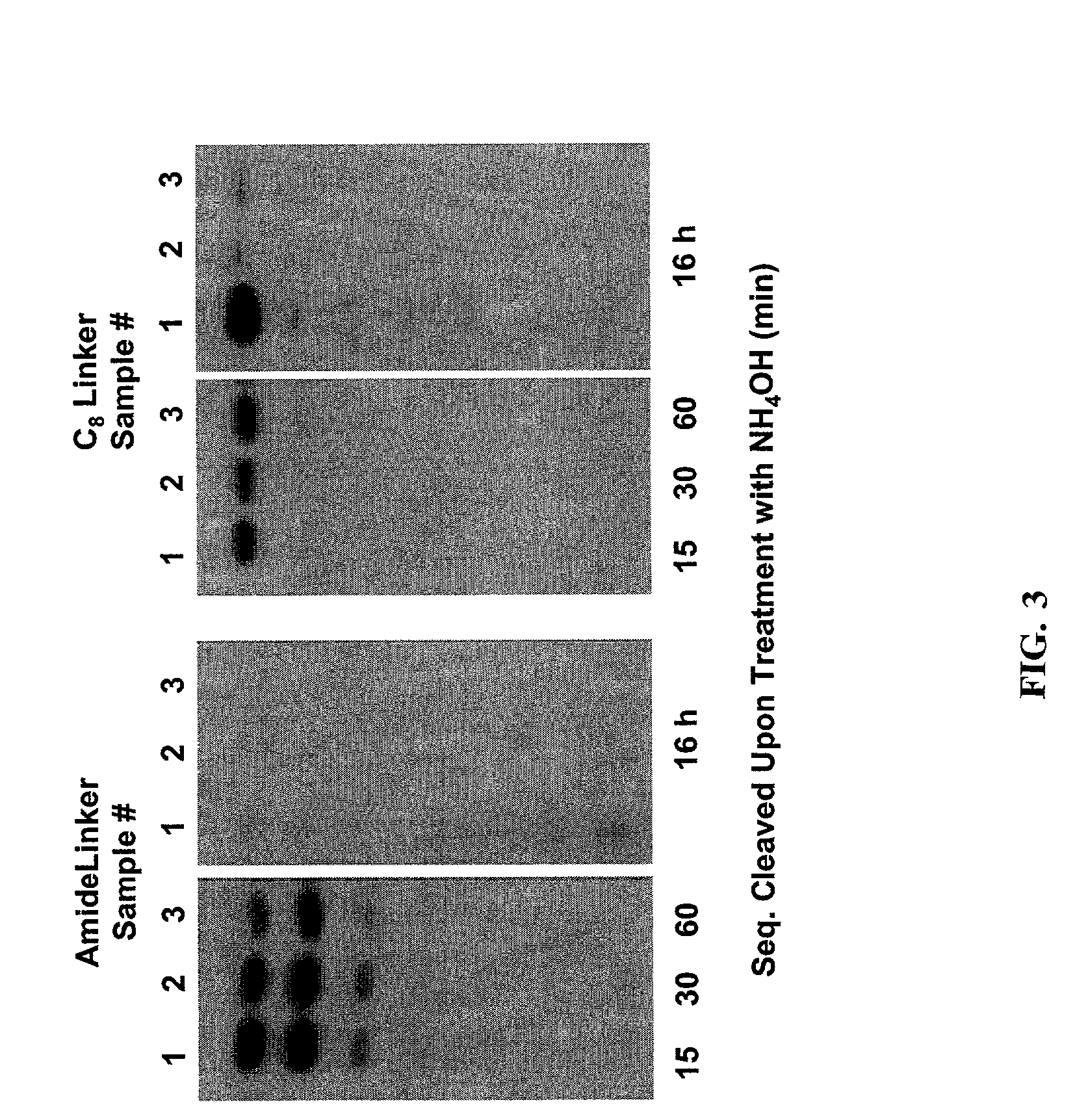
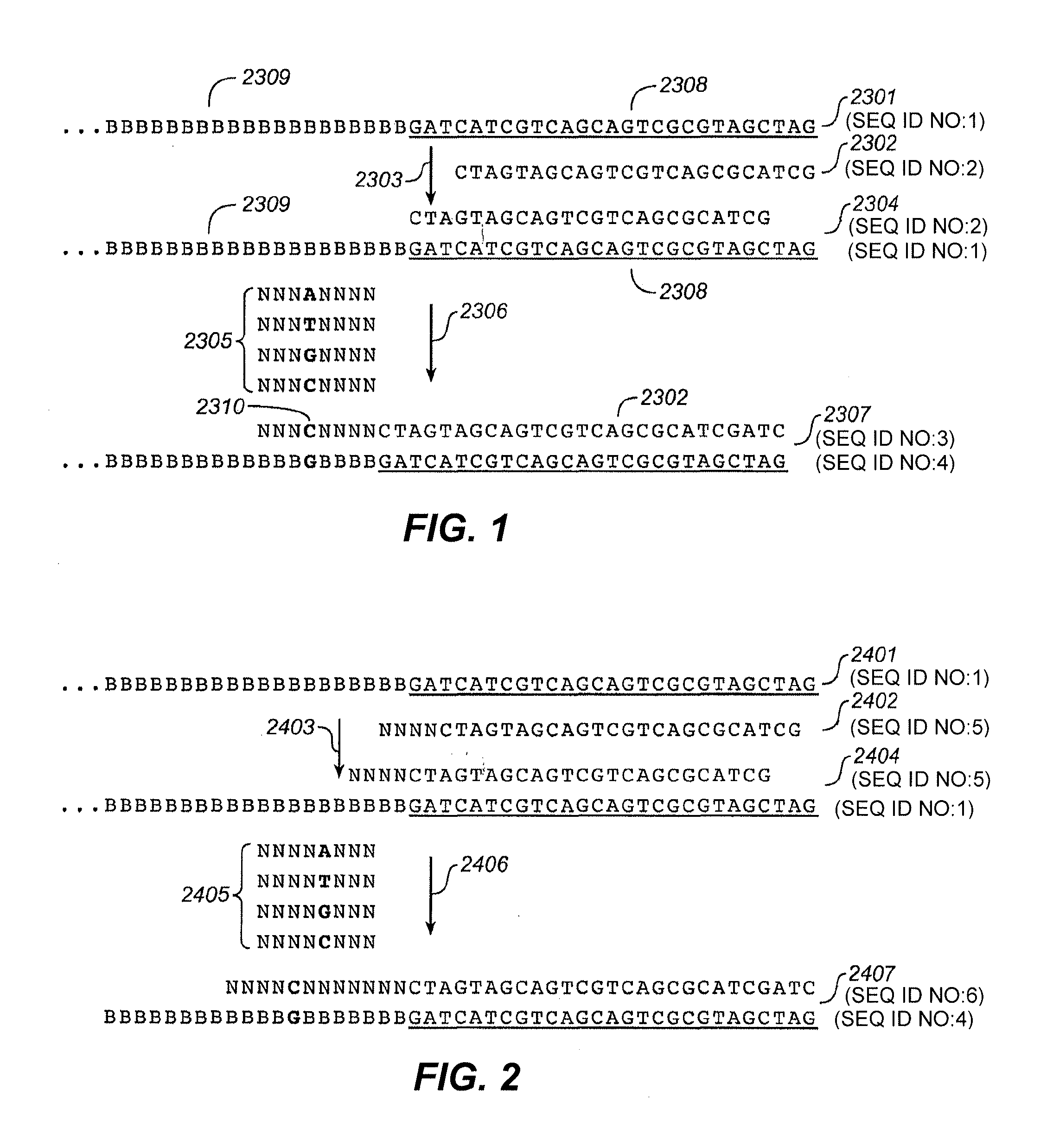
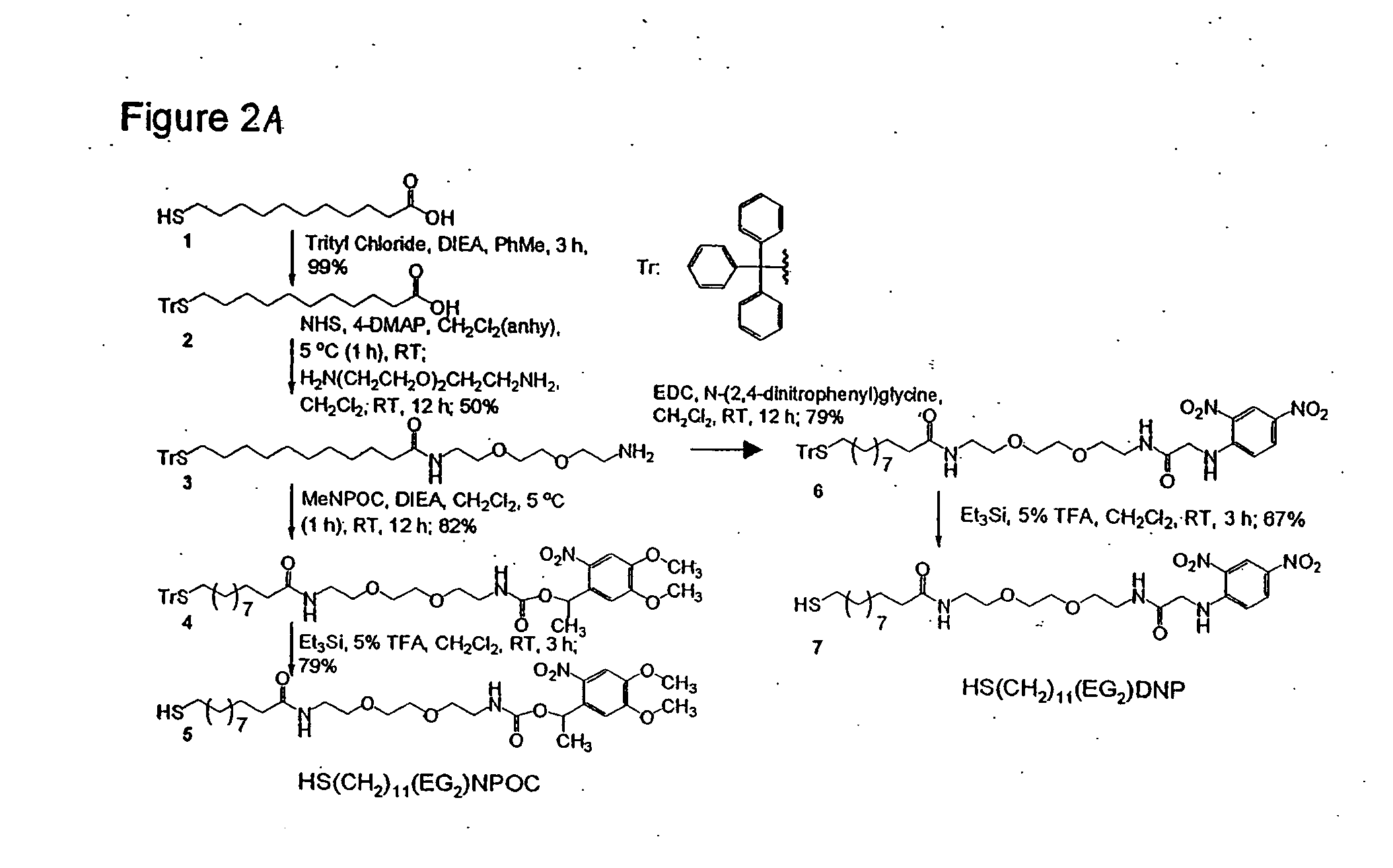
Sequencing joint and preparation method and application thereof in ultra-low frequency mutation detection
ActiveCN105861710AImprove detection accuracyNucleotide librariesMicrobiological testing/measurementMutation detectionDouble strand
The invention provides a sequencing joint and a preparation method and application thereof in ultra-low frequency mutation detection. The sequencing joint comprises a library amplification primer sequence, a target fragment amplification primer sequence and an error prompting sequence which are connected sequentially, the error prompting sequence is close to one side of a target fragment, the library amplification primer sequence is located on one side away from the target fragment, and the error prompting sequence is the sequence of a known base sequence. The error prompting sequence of the known base sequence is added on one side close to the target fragment, so that the error prompting sequence can add a specific foreign marker on each double-strand DNA template. Following sequencing data of the target fragment can be obtained conveniently, mutation introduced in the sequencing or library amplification step is screened or removed according to the fact whether the sequencing sequences have same error prompting sequences, then the sites with variation in the same positions of the two chains are determined to be real mutation, and the site with the mutation on one chain is identified as the amplification or sequencing error, so that the mutation detection efficiency is improved.
Owner:BEIJING KEXUN BIOTECH CO LTD
Biomolecule immobilization on surface via hydrophobic interactions
InactiveUS20080268440A1Bioreactor/fermenter combinationsSequential/parallel process reactionsPolystyrenePolynucleotide
A method, apparatus, or system for generating a pattern of polynucleotides on a substrate. The method includes providing a substrate having a hydrophobic surface. The method further includes conjugating a polystyrene moiety to a polynucleotide and applying a polystyrene-polynucleotide conjugate to create a plurality of reaction spots on the hydrophobic surface of the substrate. An apparatus includes a substrate with at least one polystyrene-polynucleotide conjugate on a surface of the substrate. A system can analyze a polystyrene-polynucleotide conjugate and the system may perform PCR.
Owner:APPL BIOSYSTEMS INC
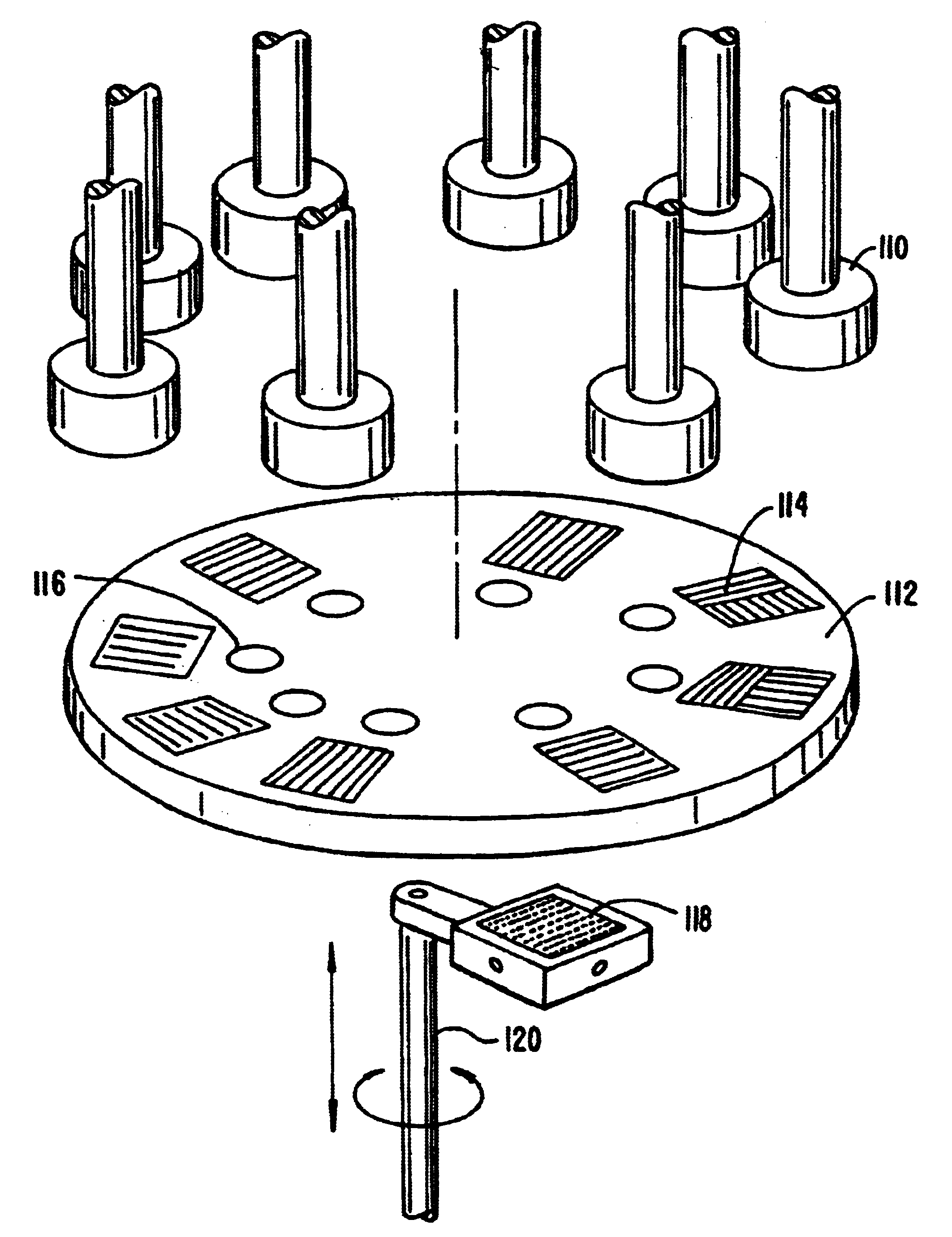
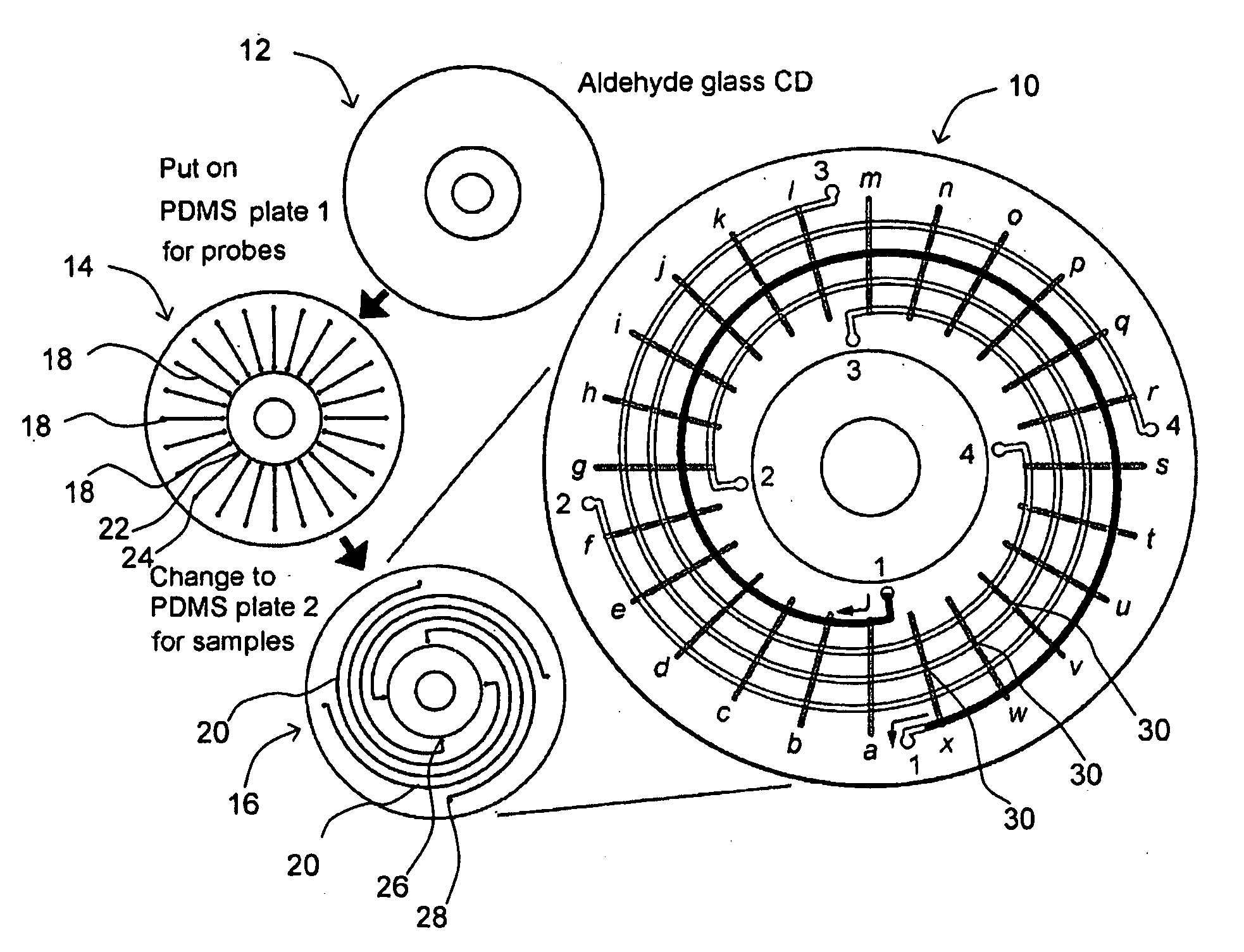
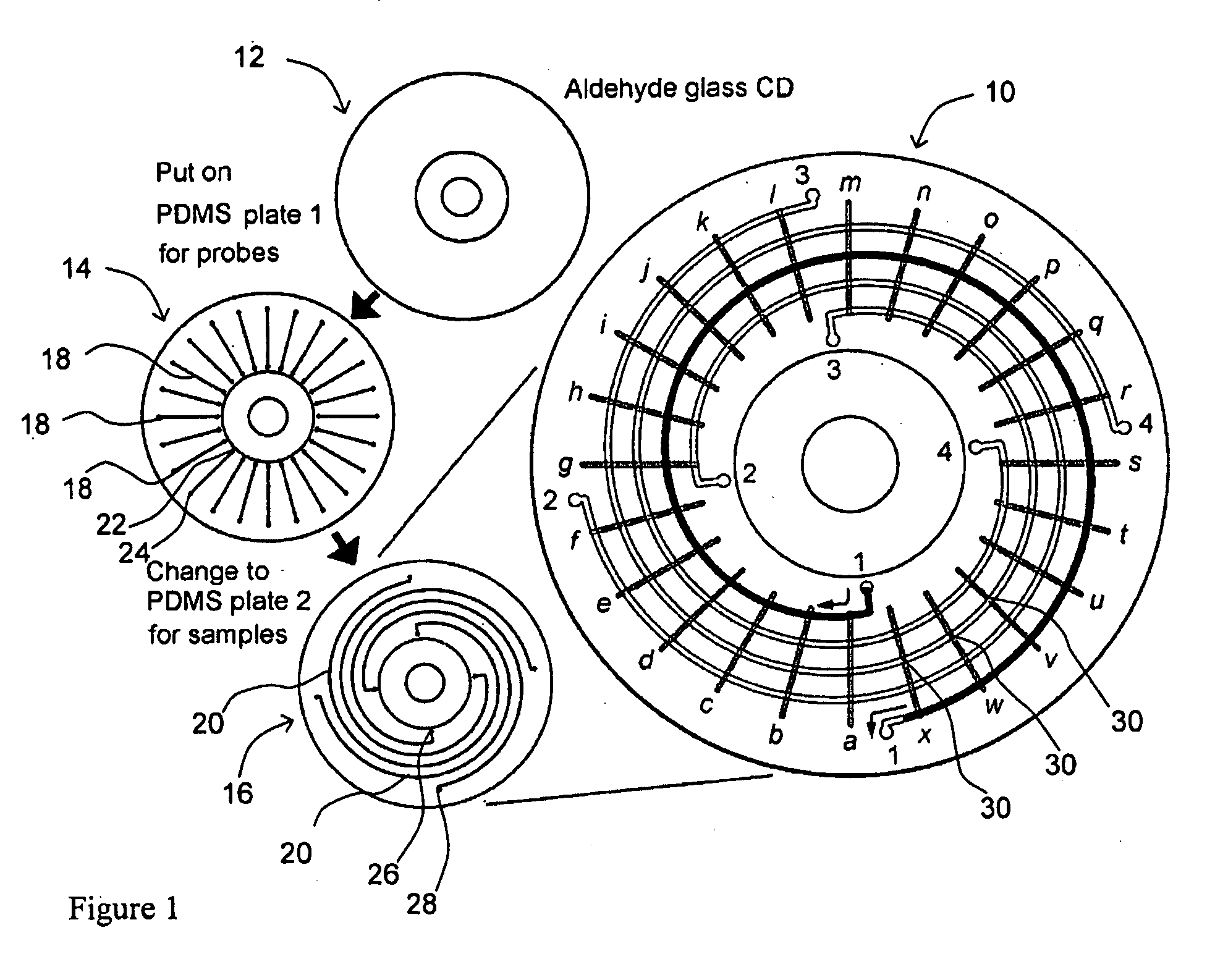
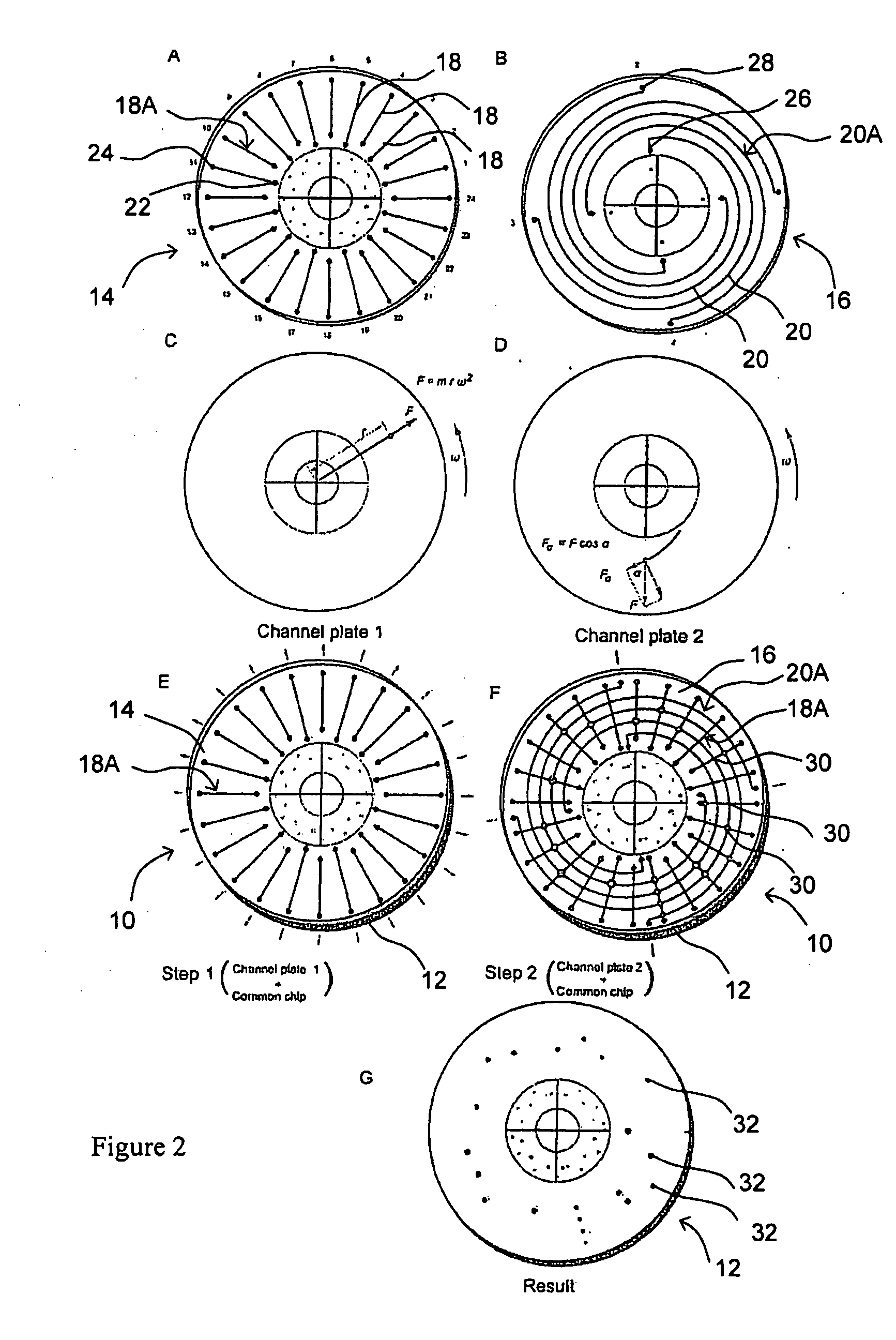
Microfluidic microarray assemblies and methods of manufacturing and using
InactiveUS20100041562A1Easy loadingImmobilised enzymesSequential/parallel process reactionsHigh densityTest sample
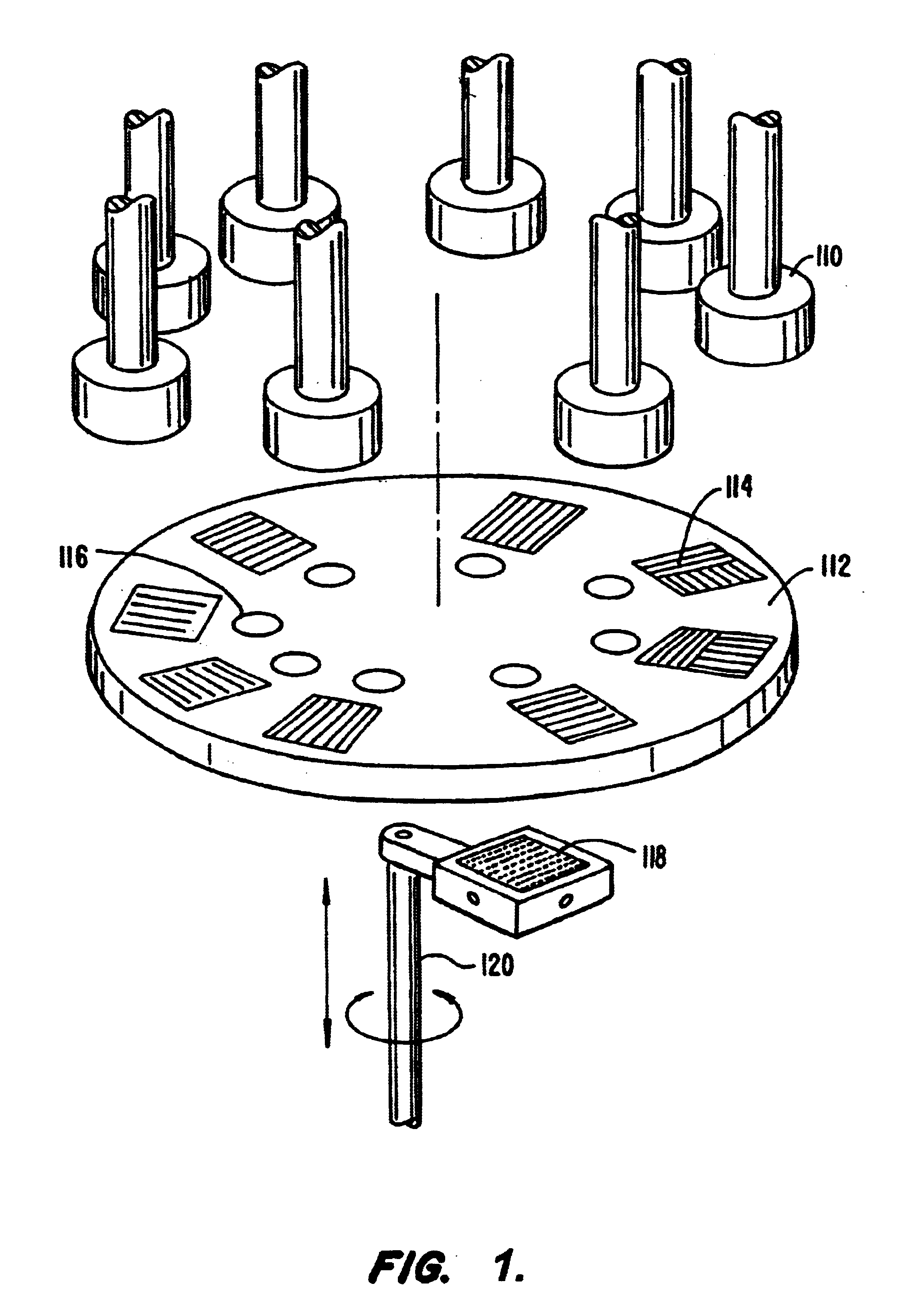
Microarray devices fabricated using microfluidic reagent distribution techniques are provided. As described herein, the invention encompasses microfluidic microarray assemblies (MMA) and subassemblies and methods for their manufacture and use. In one embodiment first and second channel plates are provided which may be sealed to a test chip in consecutive steps. Each channel plate includes microfluidic channels configured in a predetermined reagent distribution pattern. For example, the first channel plate may have a radial (linear) reagent distribution pattern and the second channel plate may have a spiral (curved) reagent distribution pattern, or vice versa. In one embodiment the first channel plate is connected to the test chip and at least one first reagent is distributed on the test chip in a first predetermined reagent pattern. The first reagent is then immobilized on the test chip. Next, the first channel plate is removed, the second channel plate is connected to the test chip and at least one second reagent is distributed on the test chip in a second predetermined reagent pattern. The first and second reagent patterns intersect to define a plurality of microarray test positions on the test chip. In one embodiment, the first reagent may comprise a plurality of separate probes each distributed to selected test position(s) of the microarray and the second reagent may comprise a plurality of test samples each distributed to selected test position(s) of the microarray. Positive or negative reactions between the probes (or other first reagent) and test samples (or other second reagent) may then be detected at the microarray test positions. For example, hybridization between selected nucleic acid probes and selected nucleic acid samples may be detected at particular test positions. The invention thus provides an efficient means to fabricate high density multi-probe, multi-sample microarrays. Preferably the test chips and channel plates are circular and centrifugal force is used to achieve fluid flow through the microfluidic channels, such as by rotating the MMA in a disc spinner.
Owner:SIMON FRASER UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com