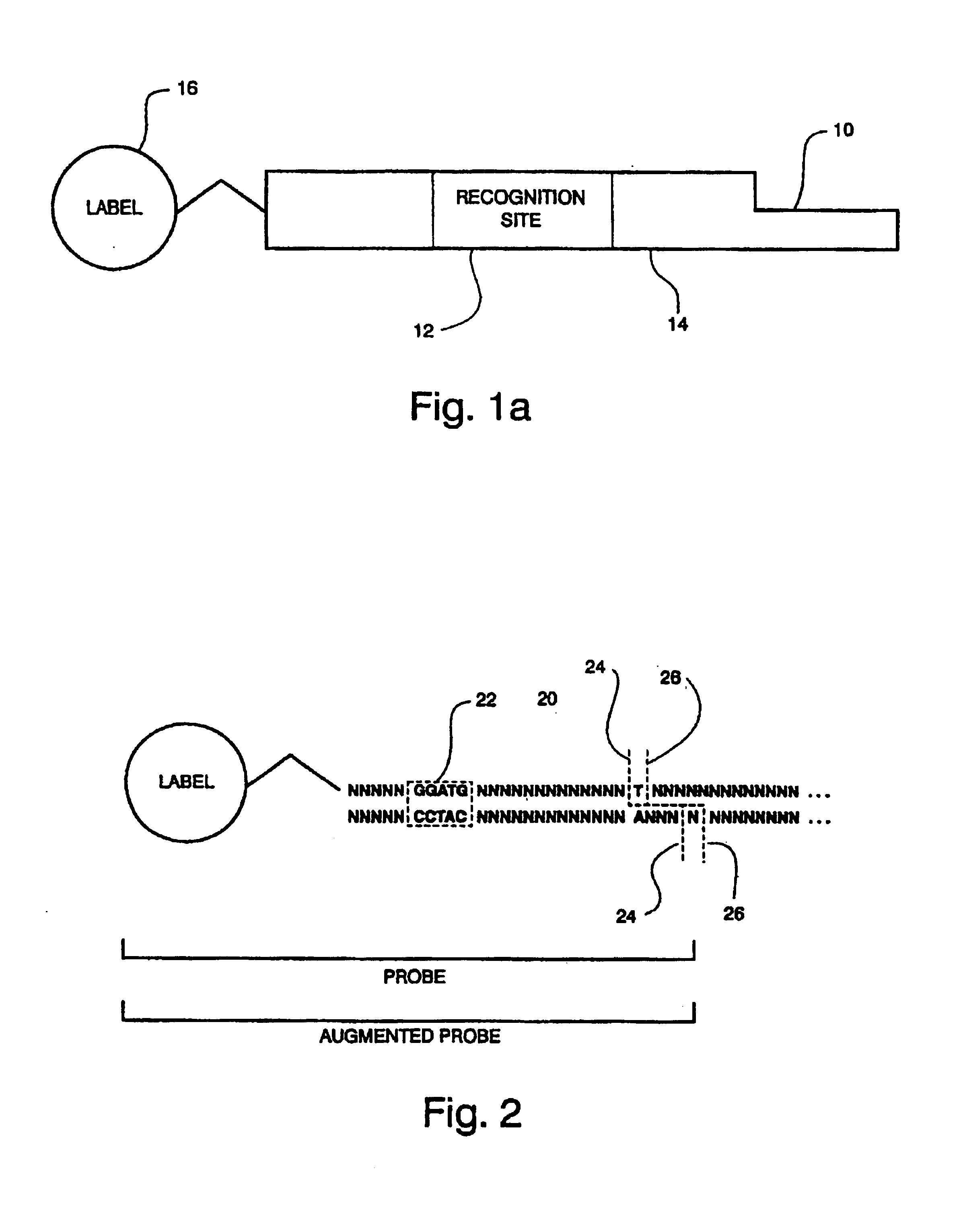
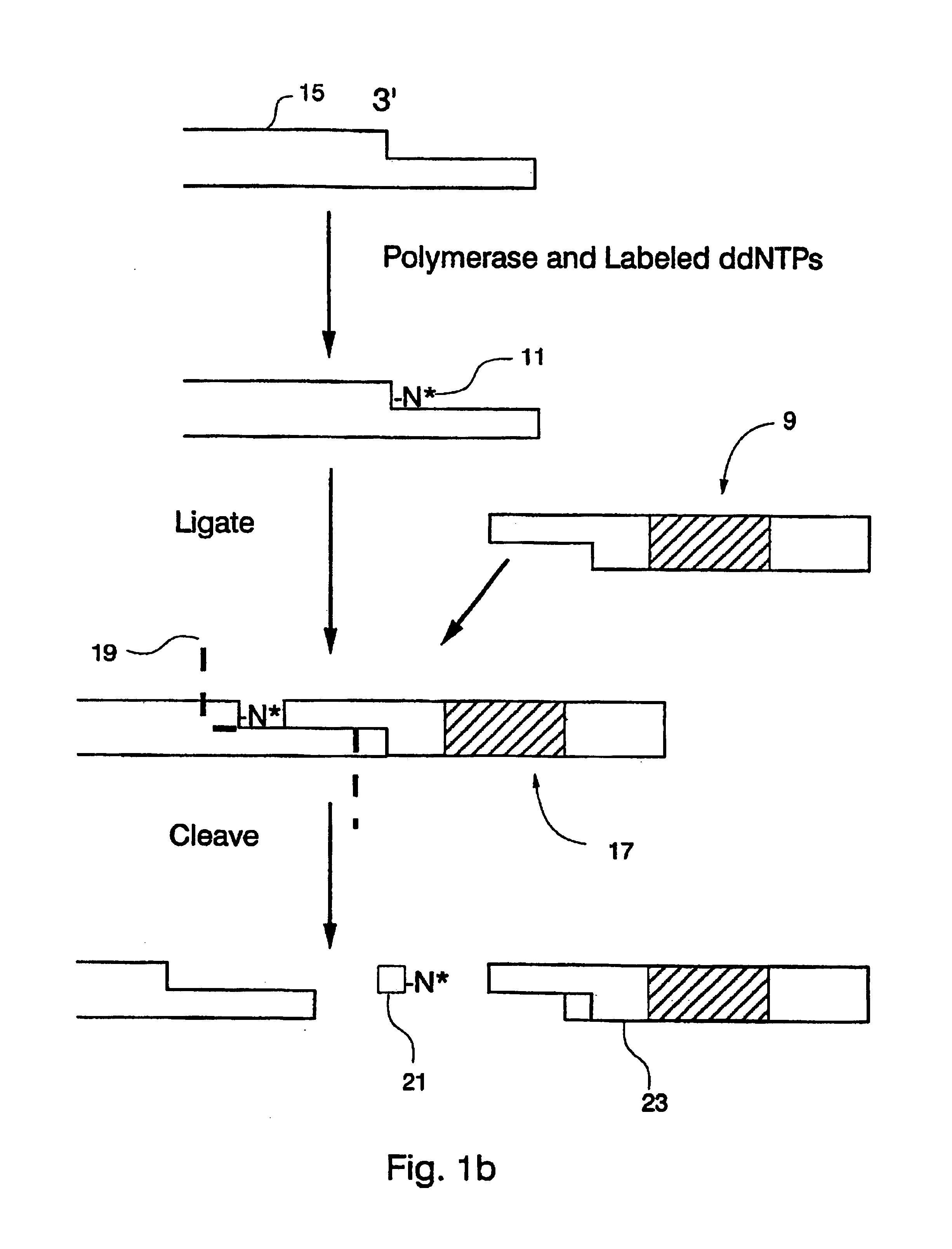
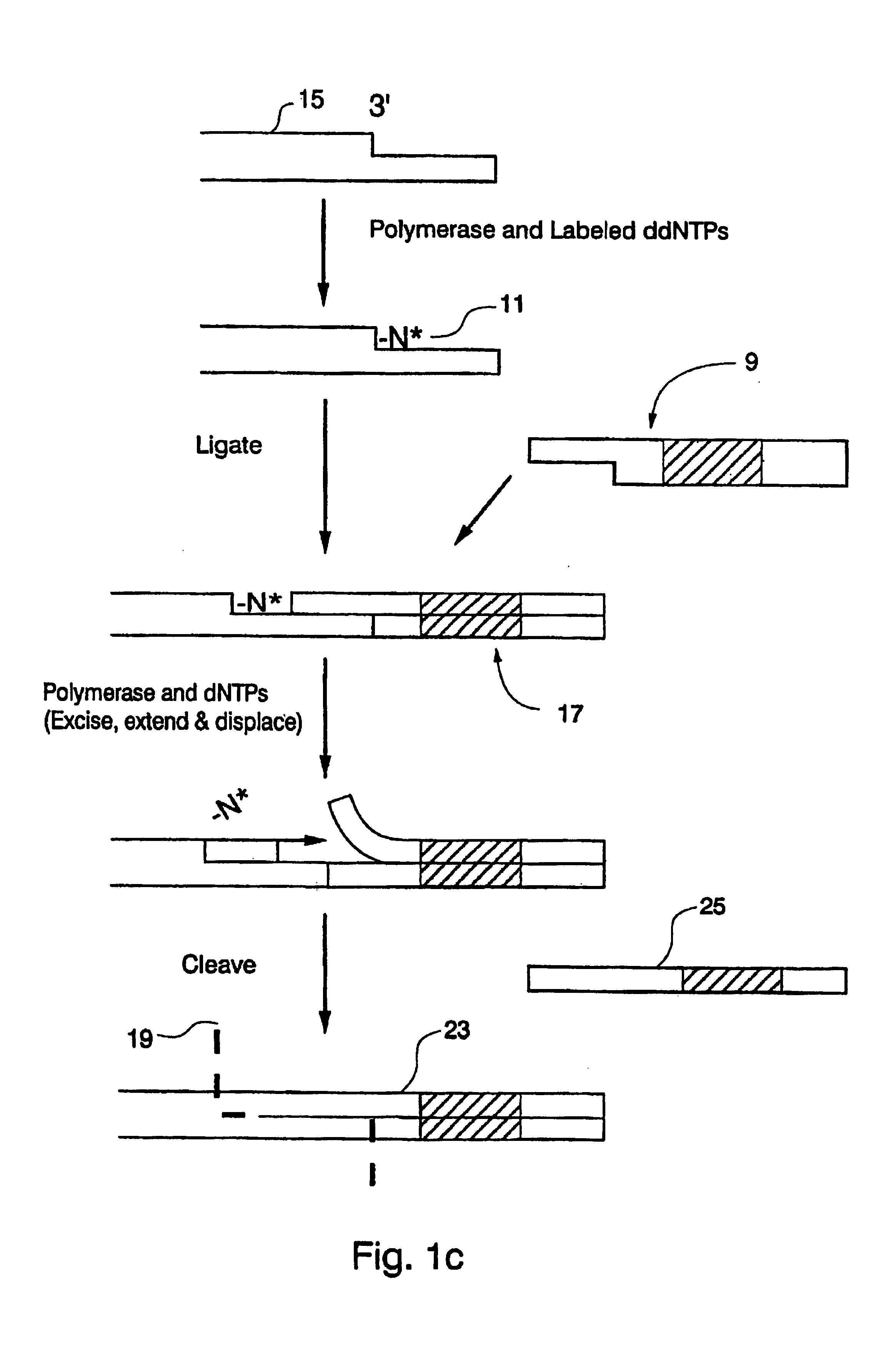
Compositions for sorting polynucleotides
a polynucleotide and compound technology, applied in the field of polynucleotides, can solve the problems of incompatibility of presence of purified signals, limited effect of such reagents, and inability to identify, sort, and/or track molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example i
Sorting Multiple Target Polynucleotides Derived from pUC 19
[0099]A mixture of three target polynucleotide-tag conjugates are obtained as follows: First, the following six oligonucleotides are synthesized and combined pairwise to form tag 1, tag 2, and tag 3 (SEQ ID NO:9, SEQ ID NO:10 and SEQ ID NO:17):
[0100]
5′-pTCGACC(w1)(w2)(w3)(w4)(w5)(w6)(w7)(w8)(w1)A GG(**)(**)(**)(**)(**)(**)(**)(**)(**)TTCGAp-5′ Tag 15′-pTCGACC(w6)(w7)(w8)(w1)(w2)(w6)(w4)(w2)(w1)A GG(**)(**)(**)(**)(**)(**)(**)(**)(**)TTCGAp-5′ Tag 25′-pTCGACC(w3)(w2)(w1)(w1)(w5)(w8)(w8)(w4)(w4)A GG(**)(**)(**)(**)(**)(**)(**)(**)(**)TTCGAp-5′ Tag 3
where “p” indicates a monophosphate, the wi's represent the subunits define in Table I, and the terms “(**)” represent their respective complements. ApUC19 is digested with Sal I and Hind III, the large fragment is purified, and separately ligated with tags 1, 2, and 3, to form pUC19-1, pUC1...
example ii
Parallel Sequencing of SV40 Fragments
[0103]A repertoire of 36-mer tags consisting of nine 4-nucleotide subunits selected from Table I is prepared by separately synthesizing tags and tag complements by a split and mix approach, as described above. The repertoire is synthesized so as to permit ligation into a Sma I / Hind III digested M13mp19. Thus, as in Example I, one set of oligonucleotides begins with the addition of A followed by nine rounds of split and mix synthesis wherein the oligonucleotide is extended subunit-wise by 3′-phosphoramidite derivatized 4-mers corresponding to the subunits of Table I. The synthesis of then completed with the nucleotide-by-nucleotide addition of one half of the Sma I recognition site (GGG), two C's, and a 5′-monophosphate, e.g. via the Phosphate-ON reagent available from Clontech Laboratories (Palo Alto, Calif.). The other set of oligonucleotides begins with the addition of three C's (portion of the Sma I recognition site) and two G's, followed by n...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Composition | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com