Double-label joint sequence for detection of tumor mutation and detection method
A linker sequence and double-label technology, which is applied in biochemical equipment and methods, microbial determination/testing, chemical library, etc., can solve the problems of inaccurate detection and inability to distinguish signal-to-noise ratio
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
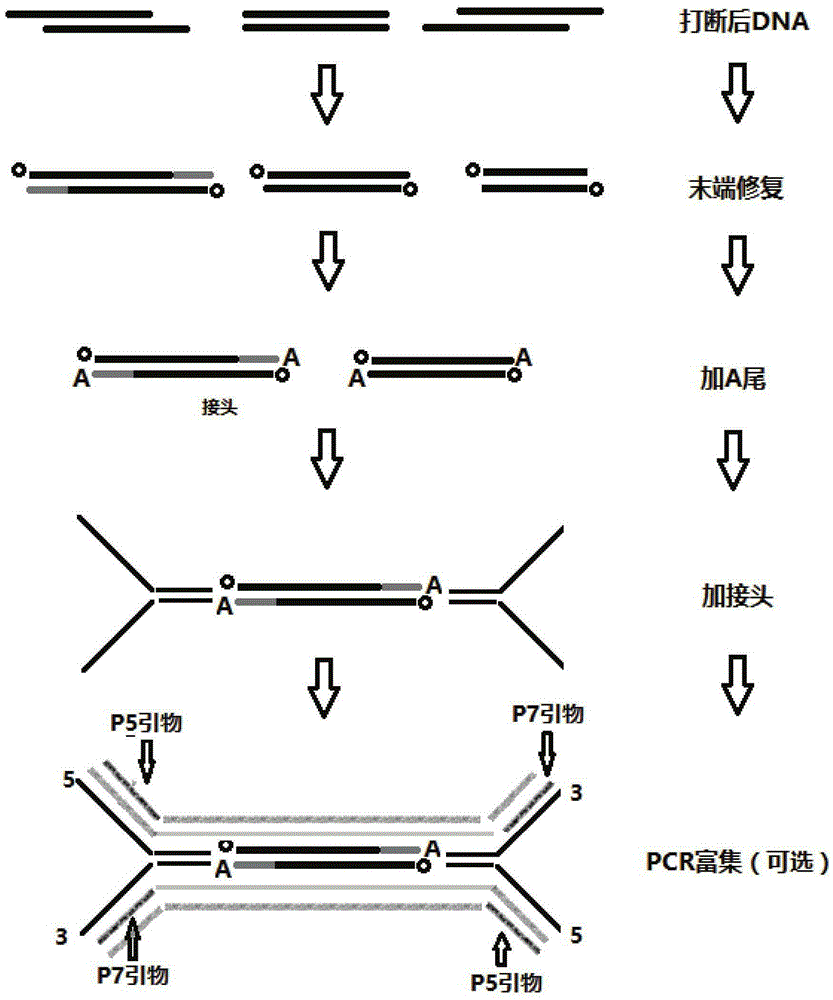
[0081] Embodiment 1: the preparation of Duplex linker
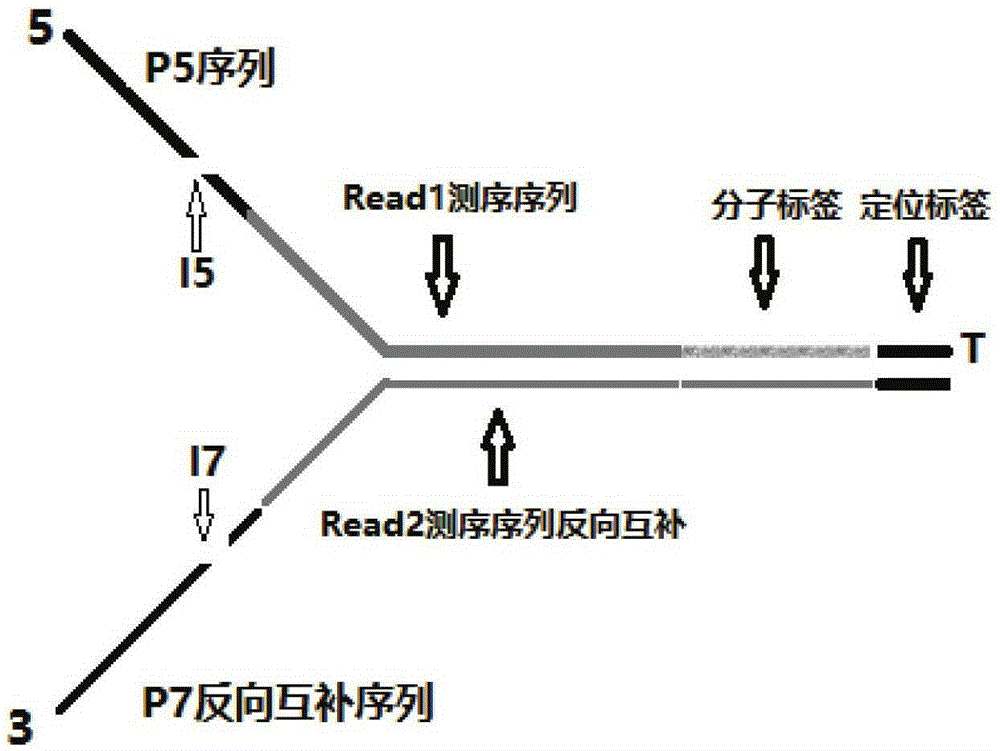
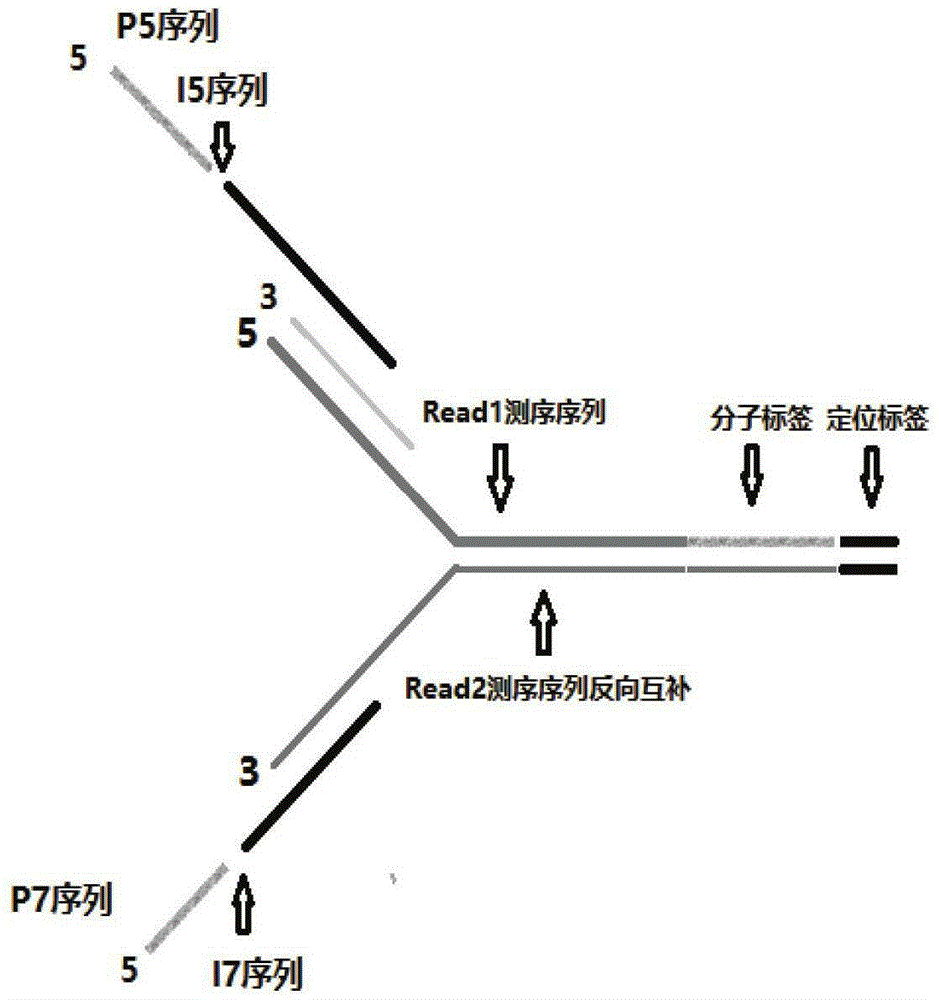
[0082] Connector primer P5, two primers of connector primer P7 (connector primer P5 is shown in the sequence obtained by connecting SEQ ID NO: 1 through the I5 index sequence of SEQ ID NO: 1; connector primer P7 is SEQ ID NO: 3 connected through the I7 index sequence SEQ ID NO: 4 is shown in the obtained sequence; wherein FFFFFFEEEEEDDDDDNNNNNNNNNNNNN is sequentially connected to the 5' end of SEQ ID NO: 3; synthetic manufacturer: Sangon Bioengineering (Shanghai) Co., Ltd.) with ddH 2 O (or TE buffer) diluted to 100uM;
[0083] Wherein FFFFF is the protection base of the enzyme cutting site, EEEEE is the enzyme cutting site, DDDDD is the positioning tag sequence, NNNNNNNNNNNN is the random molecular tag sequence, the I5 index sequence is selected from SEQ ID NO:5-12; the I7 index The sequence is selected from SEQ ID NO: 12-23.
[0084] At the same time, FFFFF / DDDDD / EEEEE / includes but is not limited to 5 identical bases;...
Embodiment 2
[0097] Embodiment 2: detection of plasma DNA mutation rate
[0098] In this embodiment: the protective base is TCTTCT; the enzyme cutting site sequence is (The positioning base is in the box, and the restriction site and the positioning base partially overlap); the molecular label is BDHVBDHV.
[0099] I5 and I7 can be: I501-I701, I502-I702, I503-I703, I504-I704, I505-I705, I506-I706, I507-I707, I508-I708, I501-I707, I502-I708, I503-I709 , I504-I710. (The base sequence corresponding to the serial number is shown in Table 1)
[0100] Sample selection and quality control: Take 5 plasma samples of lung cancer patients, use QIAGEN plasma DNA extraction kit to extract plasma DNA, use spectrophotometer to measure DNA sample purity (requires A260 / 280 between 1.8-20); then use Qubit2 .0 Measure DNA concentration (the total amount is between 5-15ng), use D1000chip (Agilent) to detect DNA sample fragment distribution (about 160-200bp) use digital PCR (Bio-rad) to measure tumor sampl...
Embodiment 3
[0122] Example 3: Cell line mutation rate detection
[0123] Two cell line DNAs, NCI-H1650 and HCT, were selected as experimental materials. NCI-1650 cell DNA was incorporated into HCT cell DNA at a mass ratio of 10%, 1%, and 0.1%, respectively. In addition, NCI-1650 and HCT cell 100% %DNA was taken as two samples respectively, correspondingly recorded as 10%, 1%, 0.1%, NCI-1650 and HCT groups. (NCI-1650 and HCT groups are only to determine the genetic background of the cell line DNA used for mixing ratios—that is, allelic locus information, such as heterozygous homozygosity, etc., through the sequencing information of these two samples, find out some Homozygous base sites, and then pick out the sites with different bases at the same site as the statistical analysis sites for other sample groups).
[0124] After the DNA samples were fully mixed, 2ug were taken for DNA library preparation (KAPA DNA library construction kit), and the 10%, 1%, and 0.1% samples after the step of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com