Lung cancer somatic mutation detection and analysis method based on high-throughput sequencing technique
A technology of somatic mutation and analysis method, applied in the field of biological detection, can solve the problems of single detection index, ineffective presentation of data information, false positives, etc., and achieve the effect of strong flexibility and accurate and reliable analysis results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
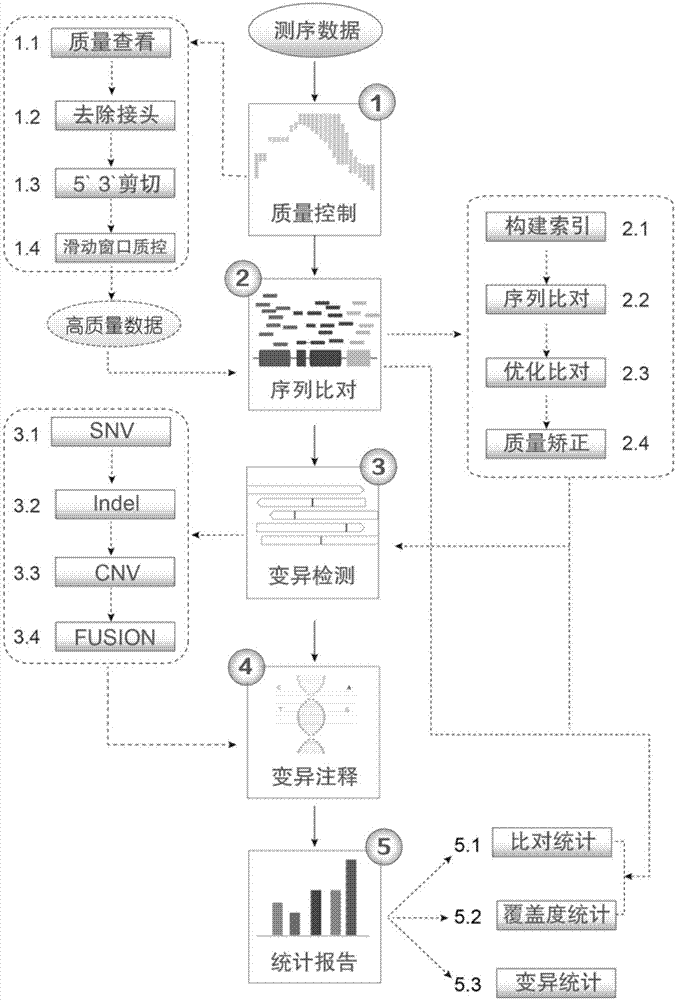
[0048] In order to achieve the purpose of the present invention, the present invention includes five main steps, (1) data quality control; (2) sequence alignment; (3) variation detection; (4) variation annotation; (5) statistical report. Such as figure 1 As shown, the specific steps and methods are as follows:
[0049] (1) Data quality control steps
[0050] (1.1) First check the sequencing data through FastQC: base quality, GC content, sequence length distribution, sequence repetition level, etc.; then use the perl language to quickly and efficiently calculate a series of quality indicators of the sequencing data, including total bases Number, number of reads, sequence length, Q20, Q30, GC content, N number and their relative percentages.
[0051] (1.2) This step uses AdapterRemoval to remove the adapter sequences contained at both ends of the sequence. This step can effectively improve the effective data and reduce subsequent alignment errors.
[0052] (1.3) In this step,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com