Cancer gene mutation and gene amplification detection
A gene and cancer technology, applied in the field of detection of genes with high incidence of cancer mutations, can solve problems such as improper primer concentration ratio, large sample consumption, and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
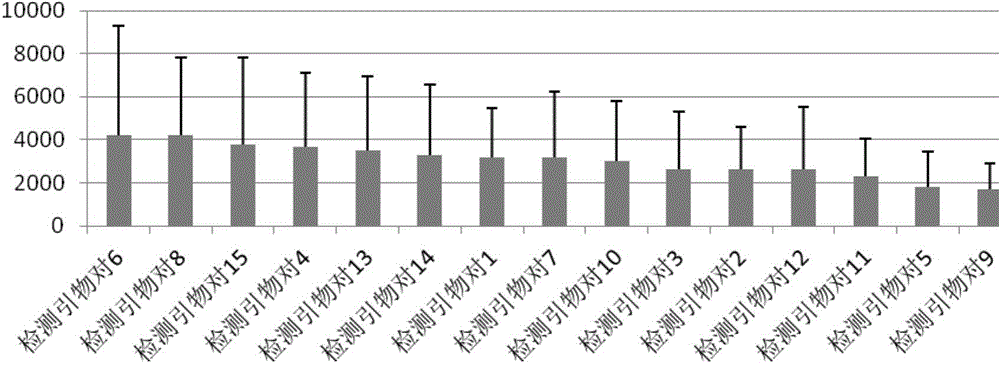
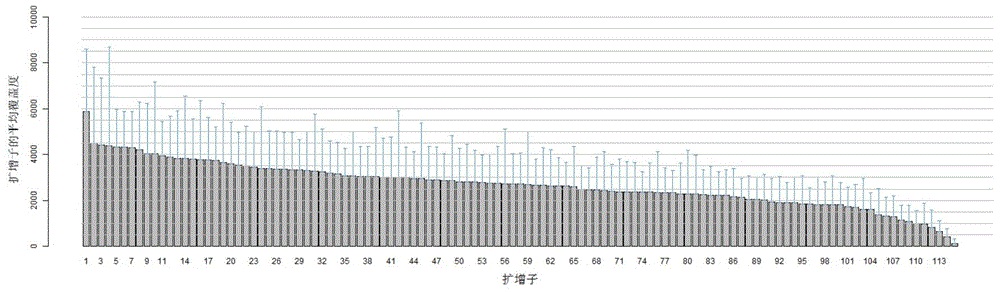
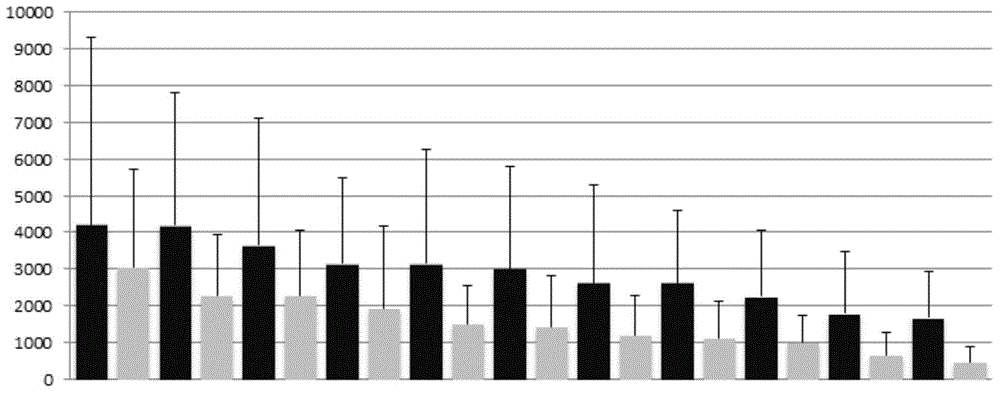
Image
Examples
Embodiment 1
[0116] Example 1: Genomic DNA is amplified using the detection primer pair of SEQ ID NO:1-30 and the balanced primer pair of SEQ ID NO:31-230
[0117] The samples used were 136 cases of lung cancer samples. The genomic DNA was subjected to multiplex PCR amplification using the detection primer pair of SEQ ID NO: 1-30 and the balanced primer pair of SEQ ID NO: 31-230, and then sequenced using the ion PGM sequencer. The specific implementation method is as follows:
[0118] a) Genomic DNA extraction
[0119] According to the standard FFEP (formalin-fixed, paraffin-embedded) tumor tissue genomic DNA extraction method, using the E.Z.N.A. FFEP DNA kit (Omega Bio-Tek), according to the manufacturer's instructions, the genome was extracted from the FFEP-treated tissue DNA. The extracted DNA was subjected to gel electrophoresis and OD value measurement for quality control and quantification with a Qubit 2.0 fluorometer (Life Technologies). The DNA concentration is required to be gr...
Embodiment 2
[0154] Example 2: DNA mutation detection and Sanger verification of various tumor samples using multiplex PCR+high-throughput sequencing technology
[0155] 1. Multiplex PCR + high-throughput sequencing to detect base mutations
[0156] Six paraffin samples were taken from 2 cases of lung cancer, 1 case of colorectal cancer, 1 case of esophageal cancer, 1 case of gastric cancer, and 1 case of ovarian cancer. For sequencing, the specific steps of multiplex PCR and high-throughput sequencing are the same as in Example 1.
[0157] The sequencing results were compared with the reference sequence to obtain the mutation detection results, as shown in Table 9.
[0158] Table 96 multiplex PCR+high-throughput sequencing base mutation detection results of samples
[0159]
[0160] 2. Sanger verification of the test results
[0161] Primers were designed for the detected sites of each of the above samples, and PCR amplification was performed respectively. The PCR products were gel-...
Embodiment 3
[0174] Example 3: Application of multiplex PCR + high-throughput sequencing technology to detect HER2 amplification mutations in tumor samples and FISH verification
[0175] 1. Multiplex PCR + high-throughput sequencing to detect HER2 amplification mutations
[0176] When the HER2 gene of the sample is amplified, the amplified copy number of the target product will increase accordingly. By calculating the ratio of the copy number of the target sequence of the HER2 gene in the sample after normalization to the corresponding copy number in the control sample, the HER2 gene in the sample can be determined. Whether there is amplification phenomenon.
[0177] Take 7 cases of esophageal cancer and 21 cases of breast cancer paraffin samples, apply detection primers to SEQ ID NO:1-30 and balance primers to SEQ ID NO:31-230 for multiplex PCR amplification, use Ion PGM sequencer for sequencing, and detect HER2 gene amplification mutation, wherein the specific steps of multiplex PCR and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com