Method and kit for analyzing ploidy of unicellular sex chromosomes quickly
A chromosome ploidy and rapid analysis technology, applied in the field of rapid analysis of single-cell sex chromosome ploidy, can solve the problems of long time, high cost, pollution, etc., and achieve the effect of improving accuracy, ensuring accuracy and reducing false positive rate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] 1. Technical principle
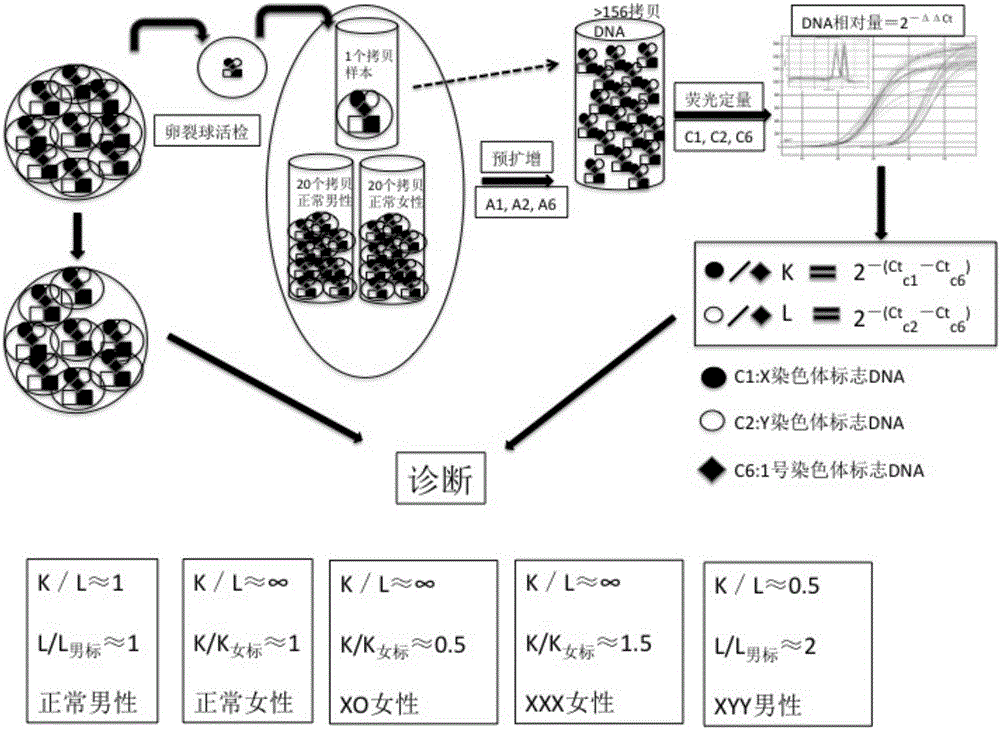
[0039] Please refer to figure 1 , figure 1 It is a schematic diagram of the technology for rapid analysis of unicellular sex chromosome ploidy in the present invention.
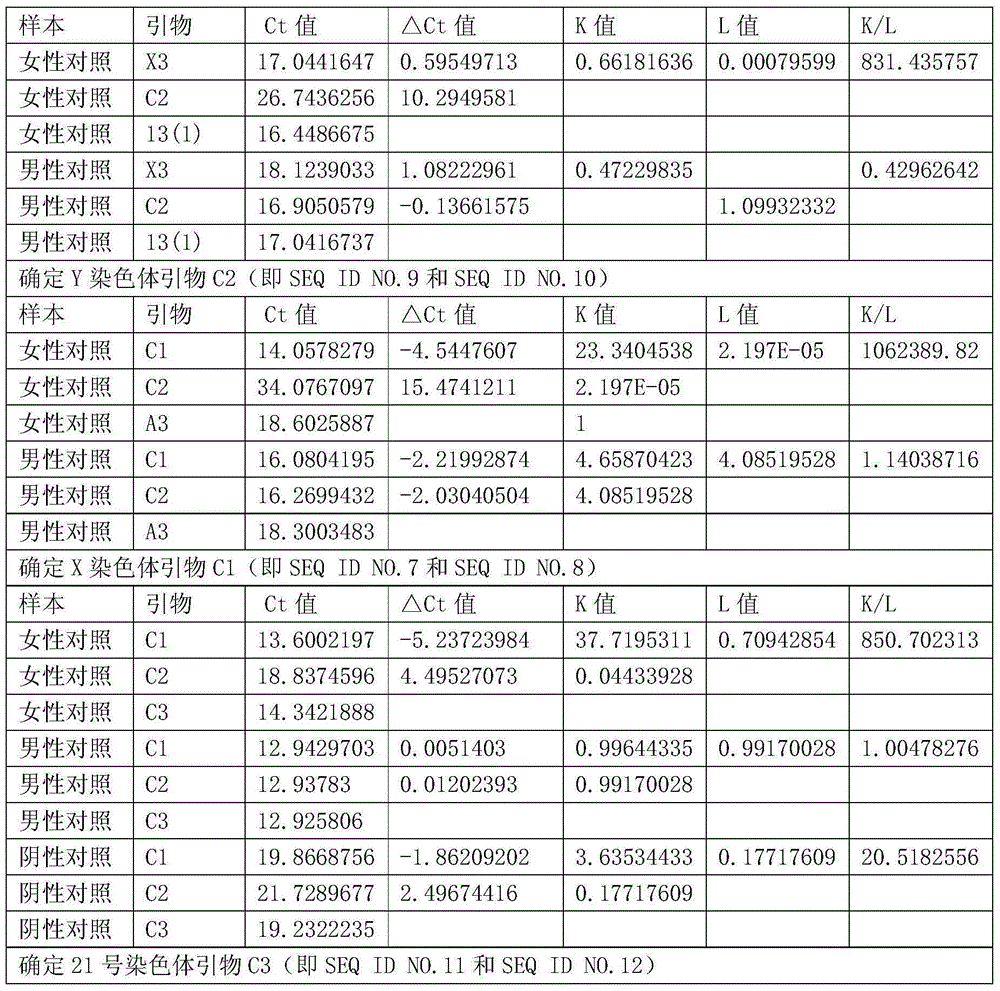
[0040] The kit of the invention is based on single cell DNA amplification and real-time quantitative PCR technology. Firstly, the trace DNA from blastomeres or embryonic trophoblast cells of embryos is amplified in equal amounts (the first round of amplification) for special fragments (marker DNA fragments on X, Y and autosomes), and then quantitatively using real-time PCR (the second round of amplification) determines the Ct value of the marker DNA fragment in each sample (the number of cycles required to reach the saturation concentration), ΔCt is the difference between the marker gene and the endogenous reference gene Ct value; ΔΔCt is the difference between the two markers The difference between gene Ct values. Since there is an inverse linear relationship between the C...
Embodiment 2
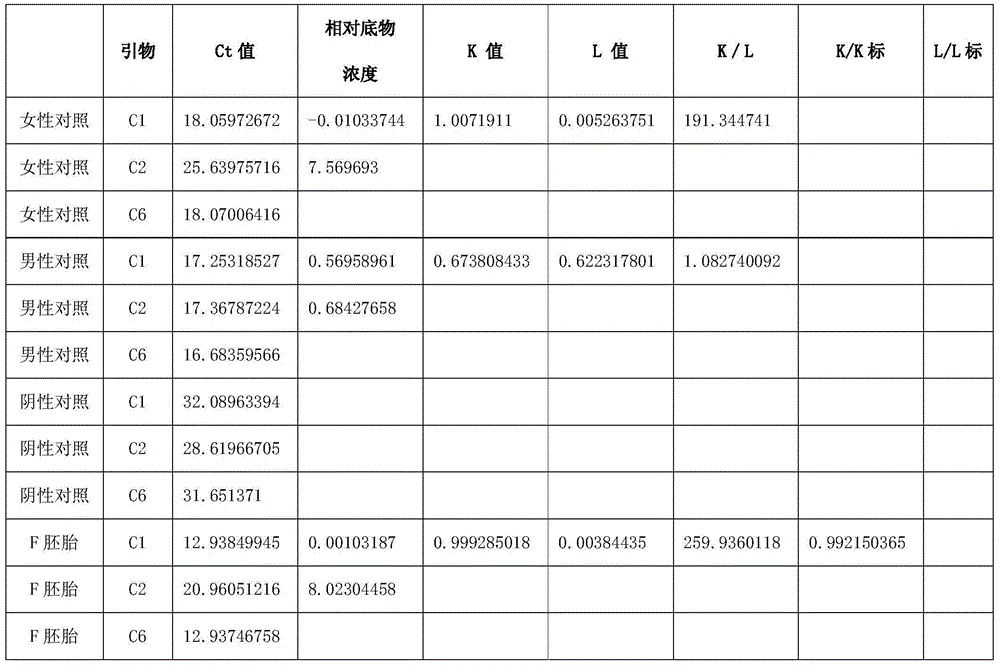
[0062] Using one case of donated embryo, analyze the ploidy of sex chromosomes. The diagnostic results are shown in Table 2, suggesting that the embryo F is female with a normal sex chromosome ratio of XX.
[0063] Table 2 Diagnosis results of unicellular sex chromosome ploidy analysis
[0064]
Embodiment 3
[0066] The accuracy of using the method of the present invention to quickly analyze the ploidy of unicellular sex chromosomes is further verified with a large sample. Collect volunteers from 100 normal males, 100 normal females, 8 cases of XXY Klinefelter syndrome, 7 cases of XO Turner syndrome, 4 cases of XYY syndrome, and 7 cases of multiple X syndrome, collect peripheral blood of volunteers, and extract whole genome DNA , using the primers of Example 1, according to the method of Example 1 to analyze unicellular sex chromosome ploidy, wherein the amount of whole genome DNA template used in the first round of PCR is 3pg. The result shows that the accuracy of the method of the present invention is 100%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com