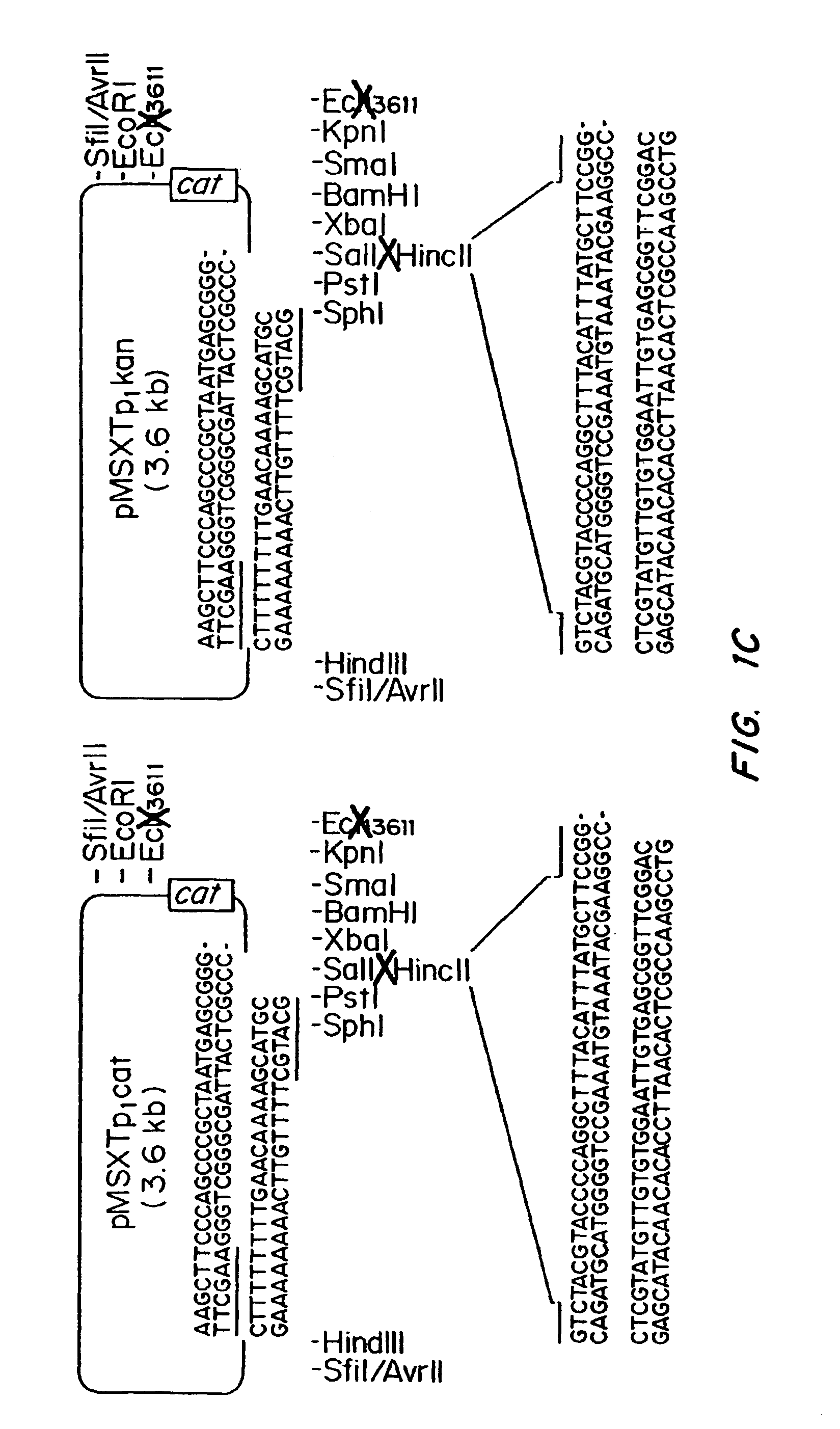
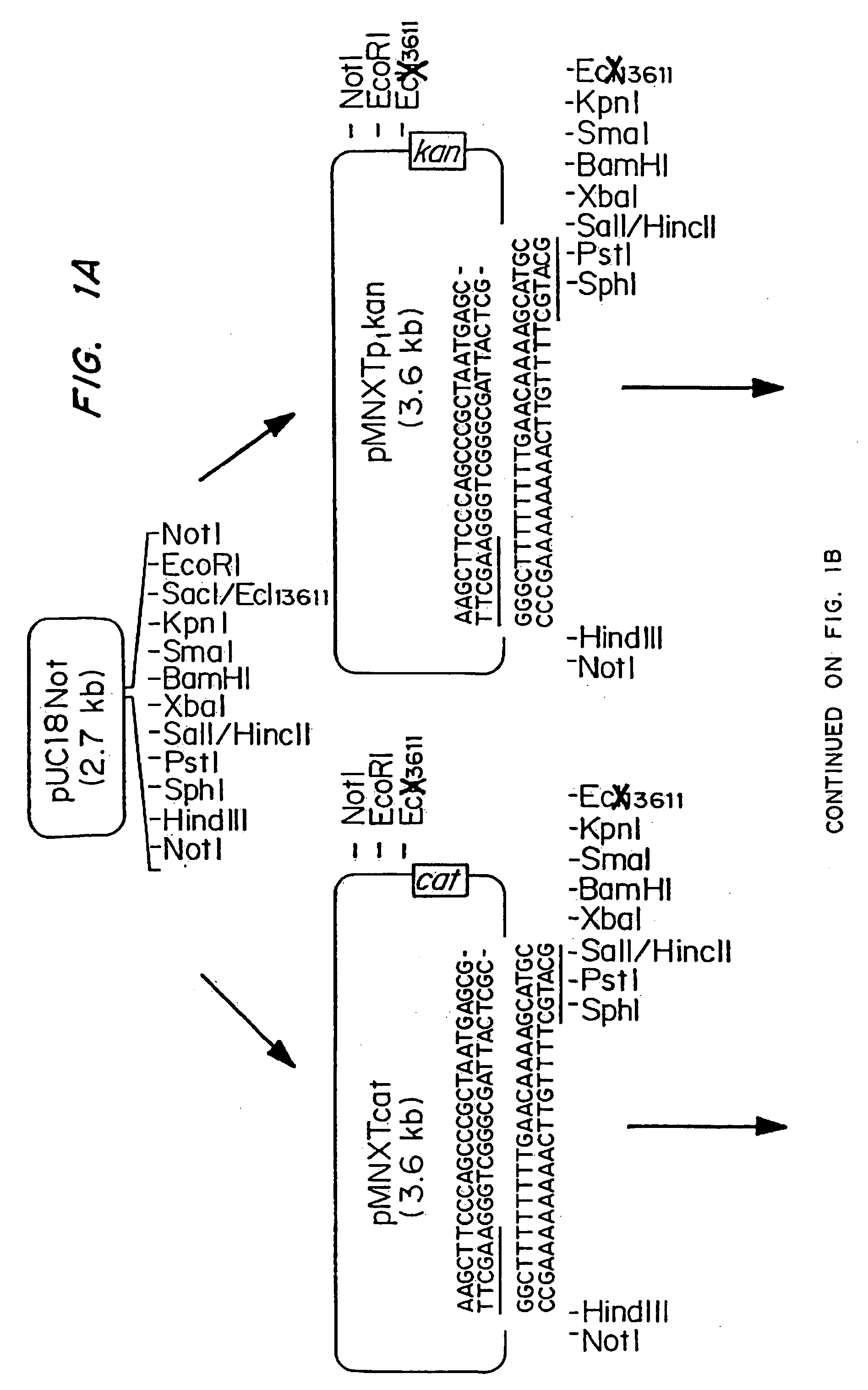
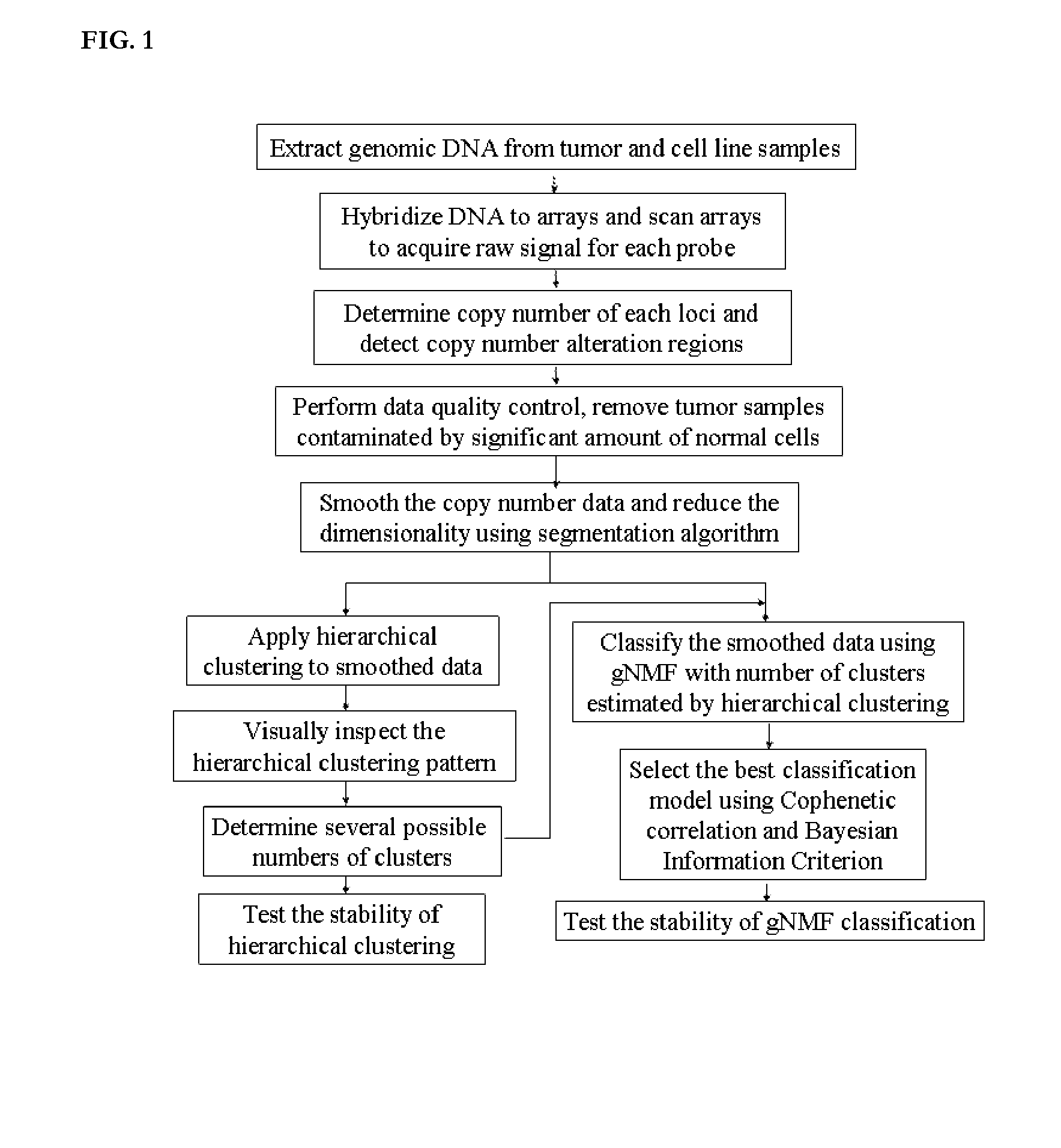
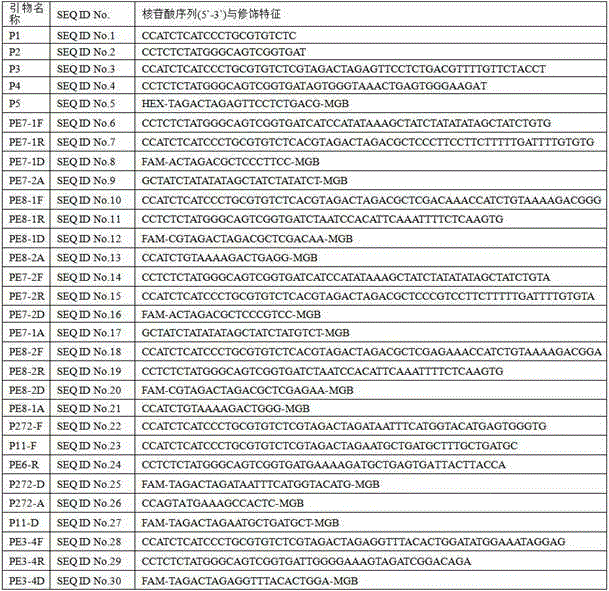
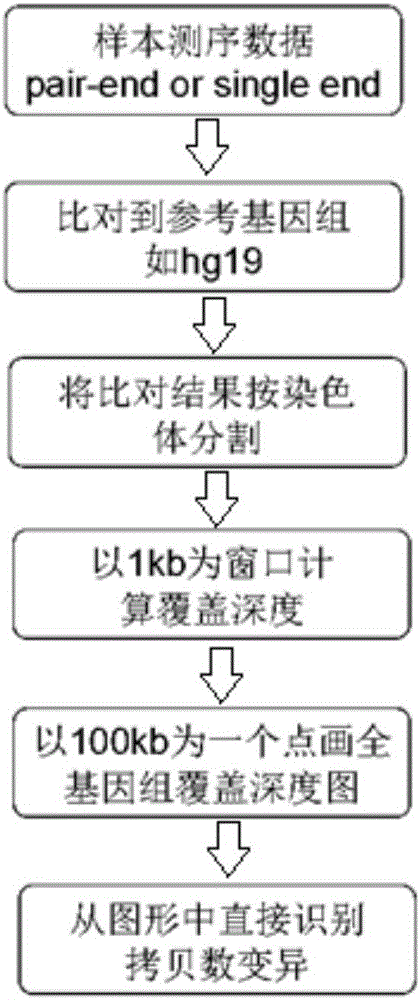
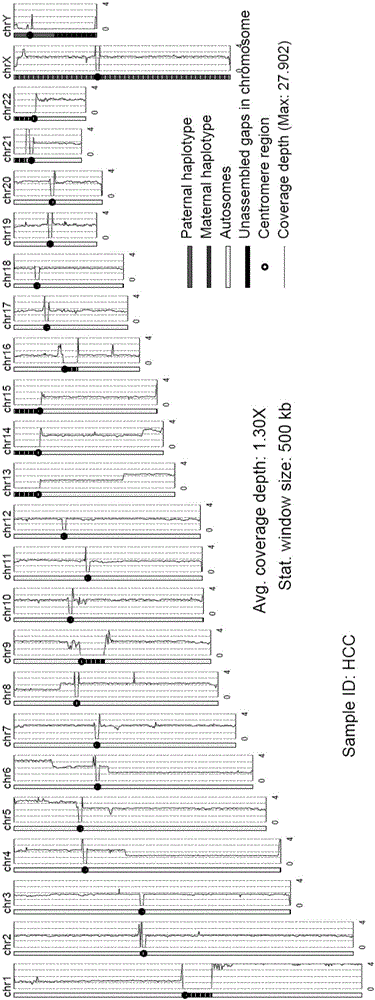
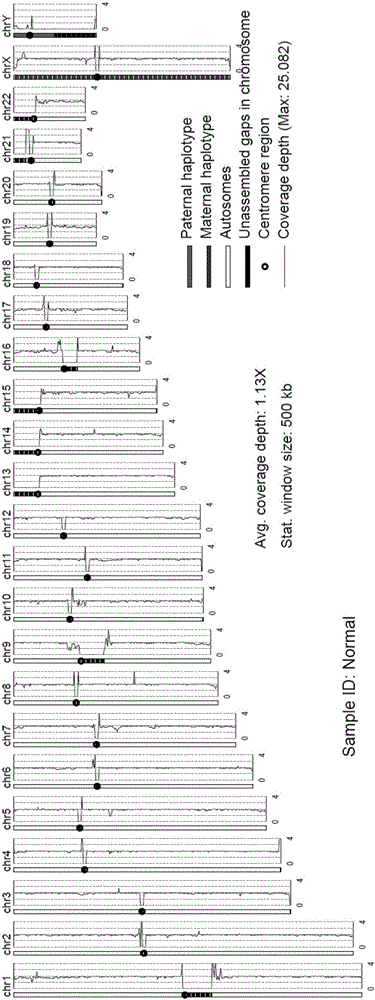
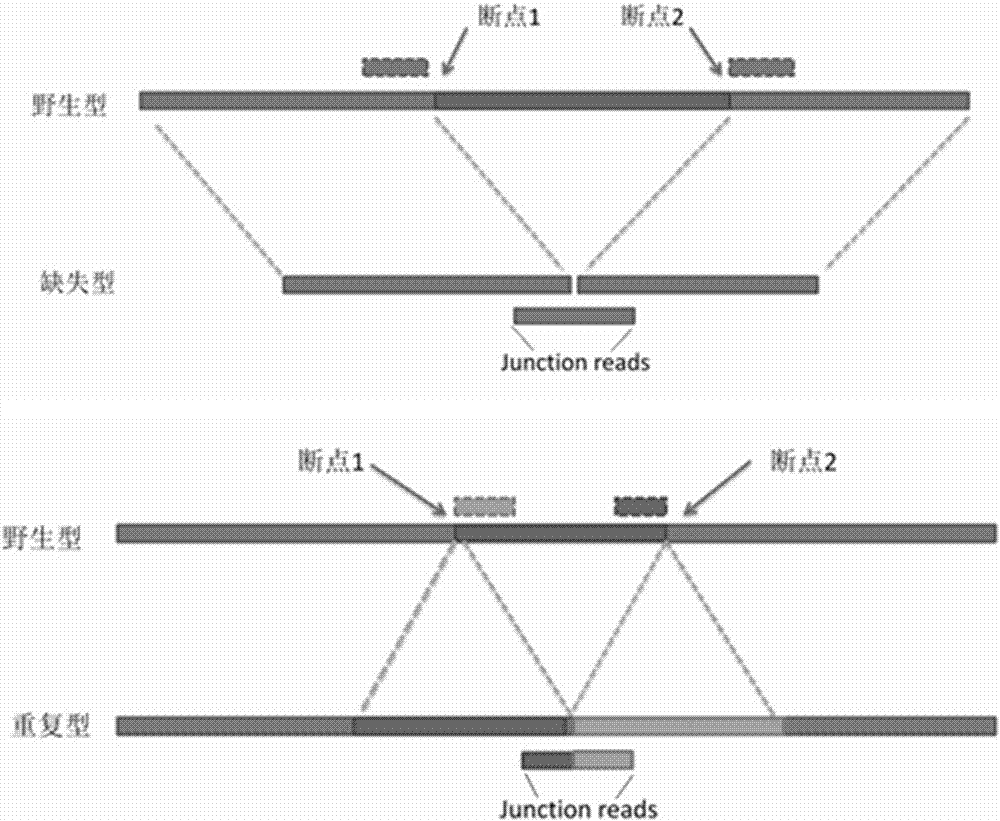
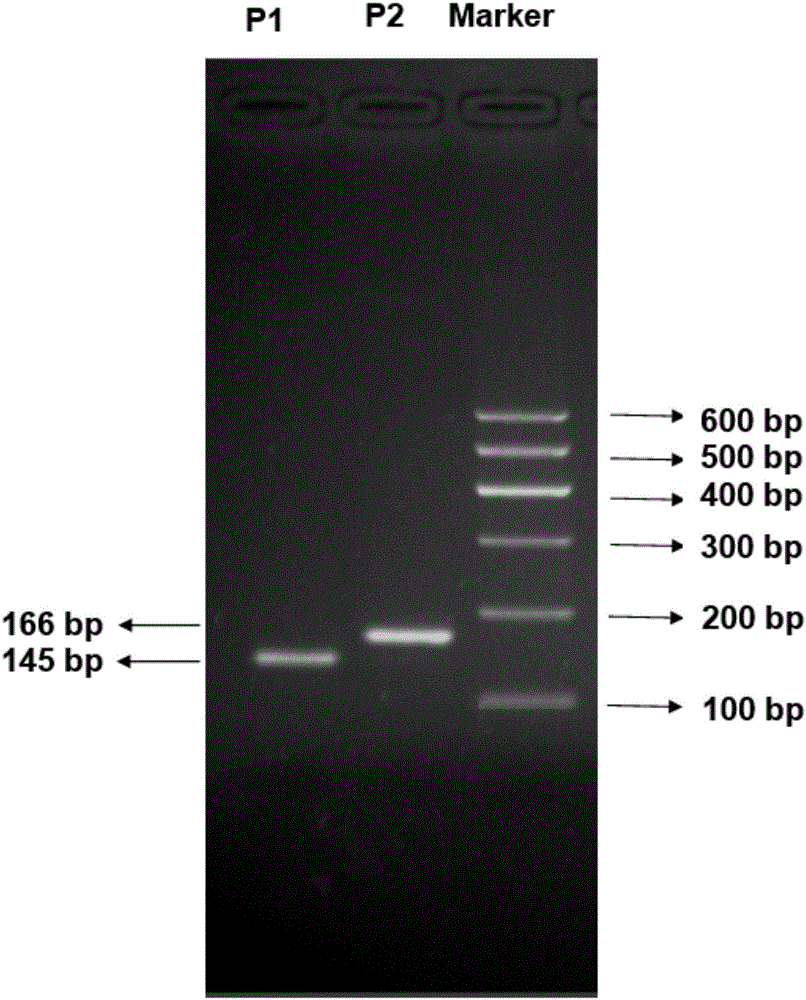
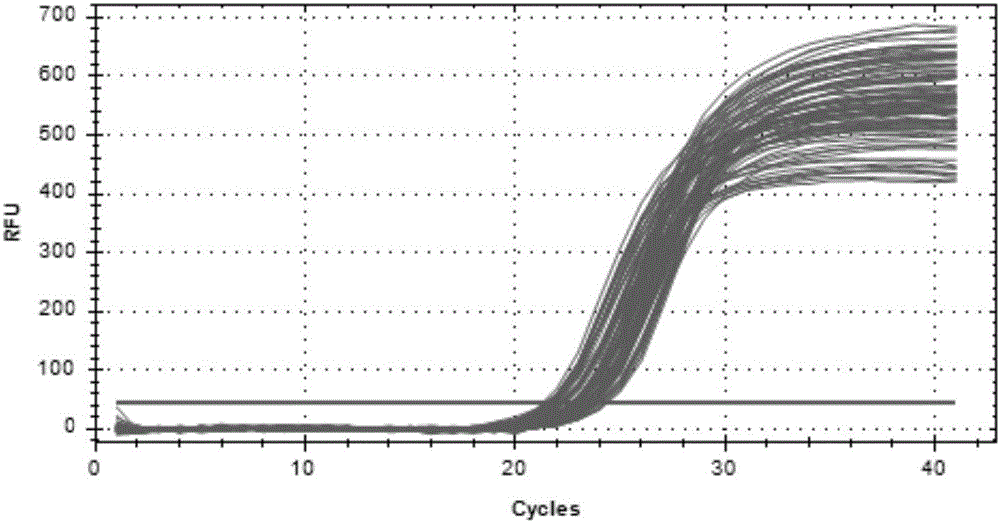
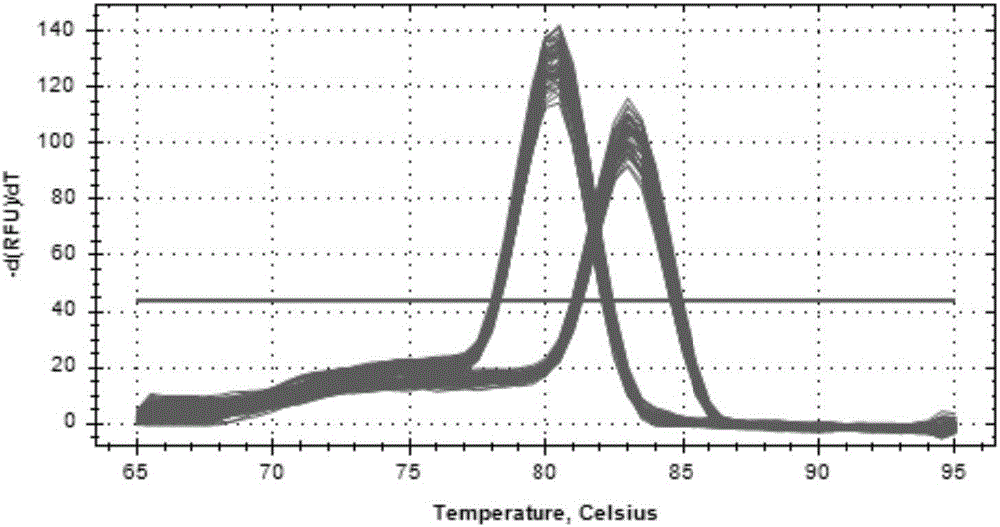
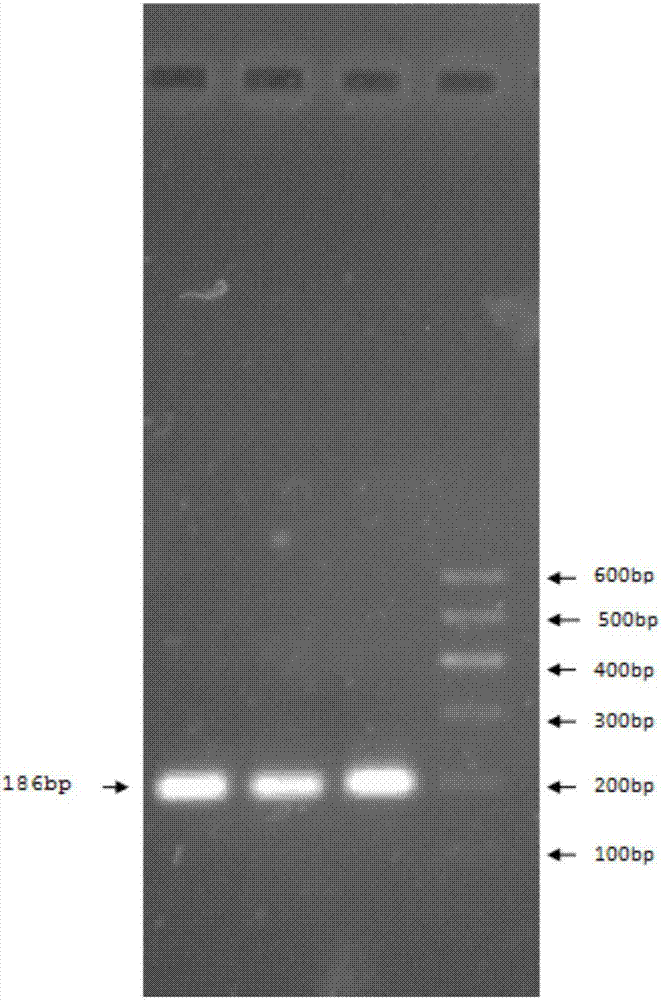
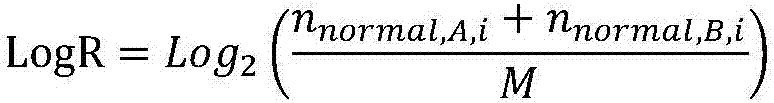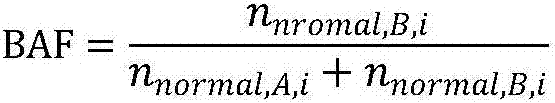Patents
Literature
342 results about "Gene copy" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Analysis of genetic polymorphisms and gene copy number
InactiveUS6468744B1Material nanotechnologySequential/parallel process reactionsCytochrome P450Drug biotransformation
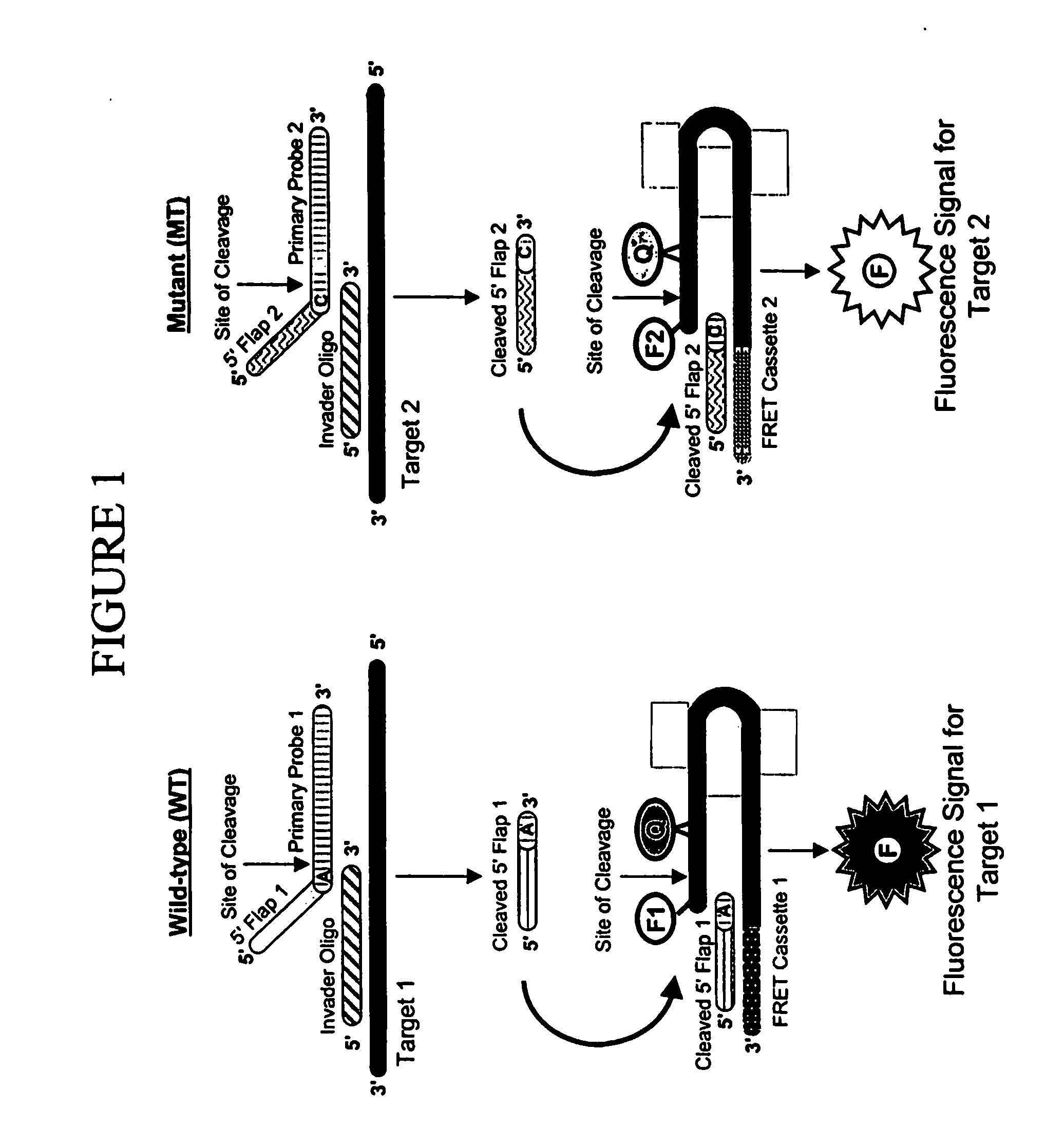
The invention provides methods for detecting variations in polymorphic sites and / or variations in gene copy number. The methods are particularly useful for analysis of biotransformation genes, such as cytochromes P450.
Owner:AFFYMETRIX INC
Compositions and methods for cancer diagnosis and therapy
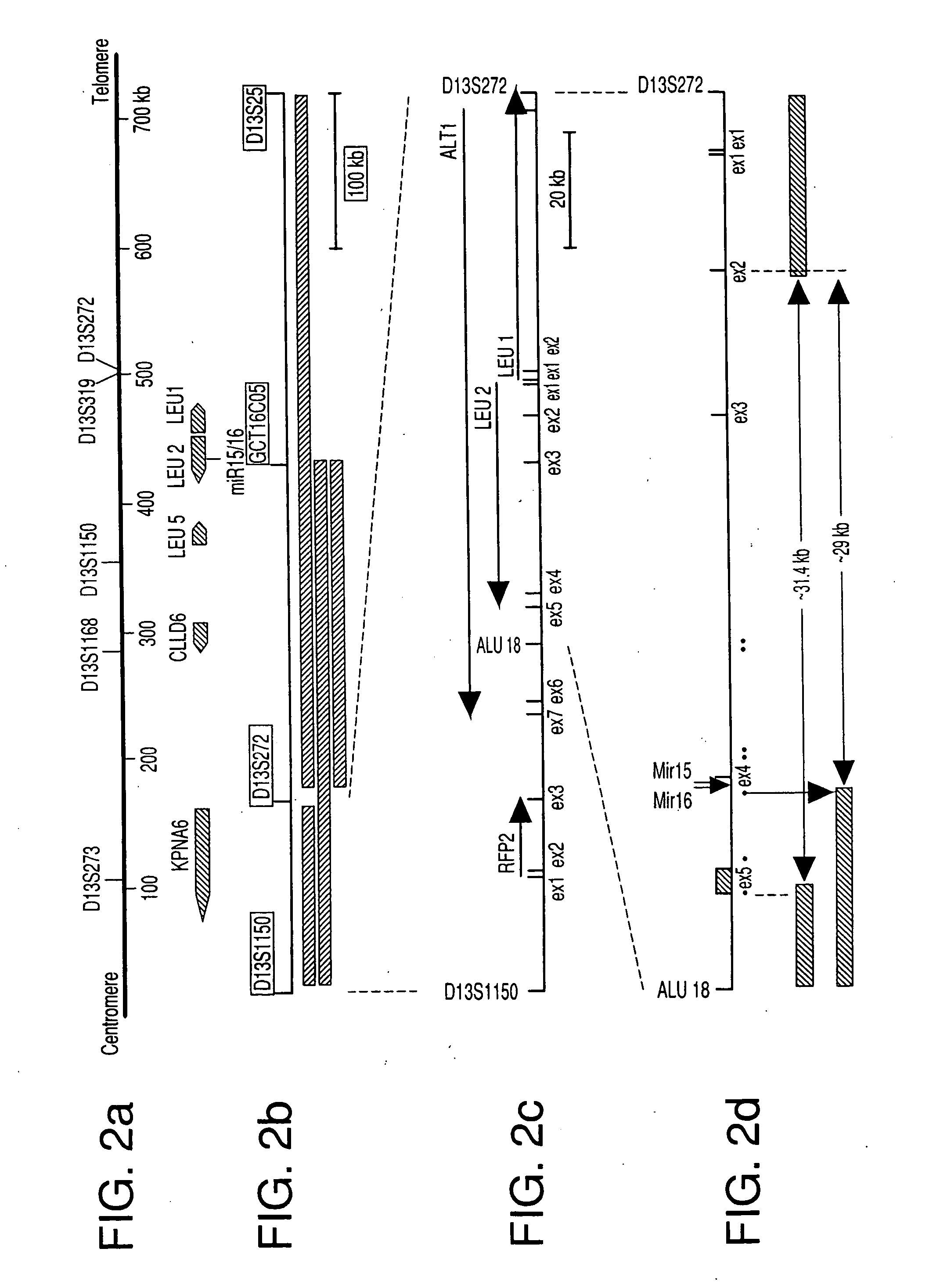
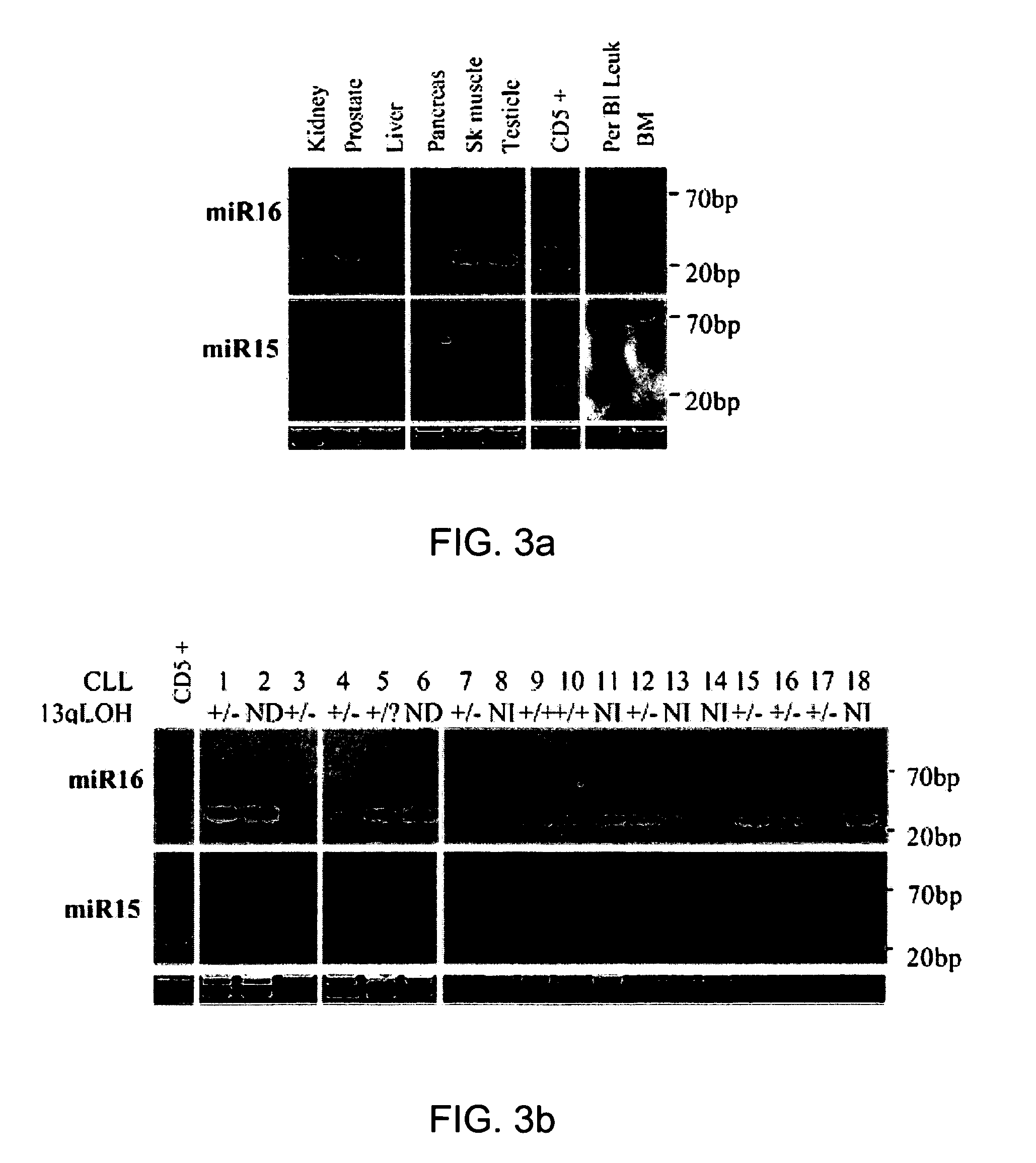
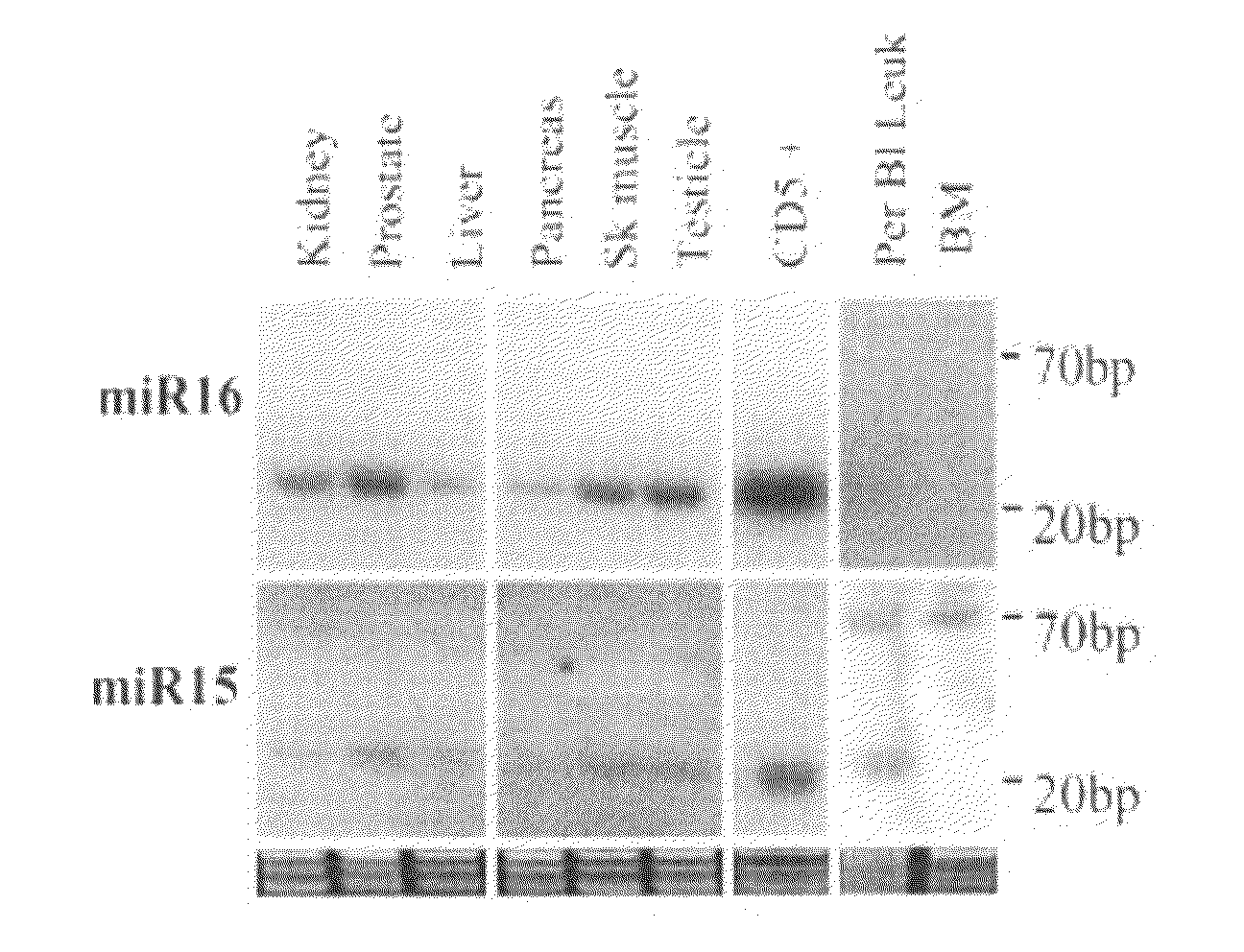
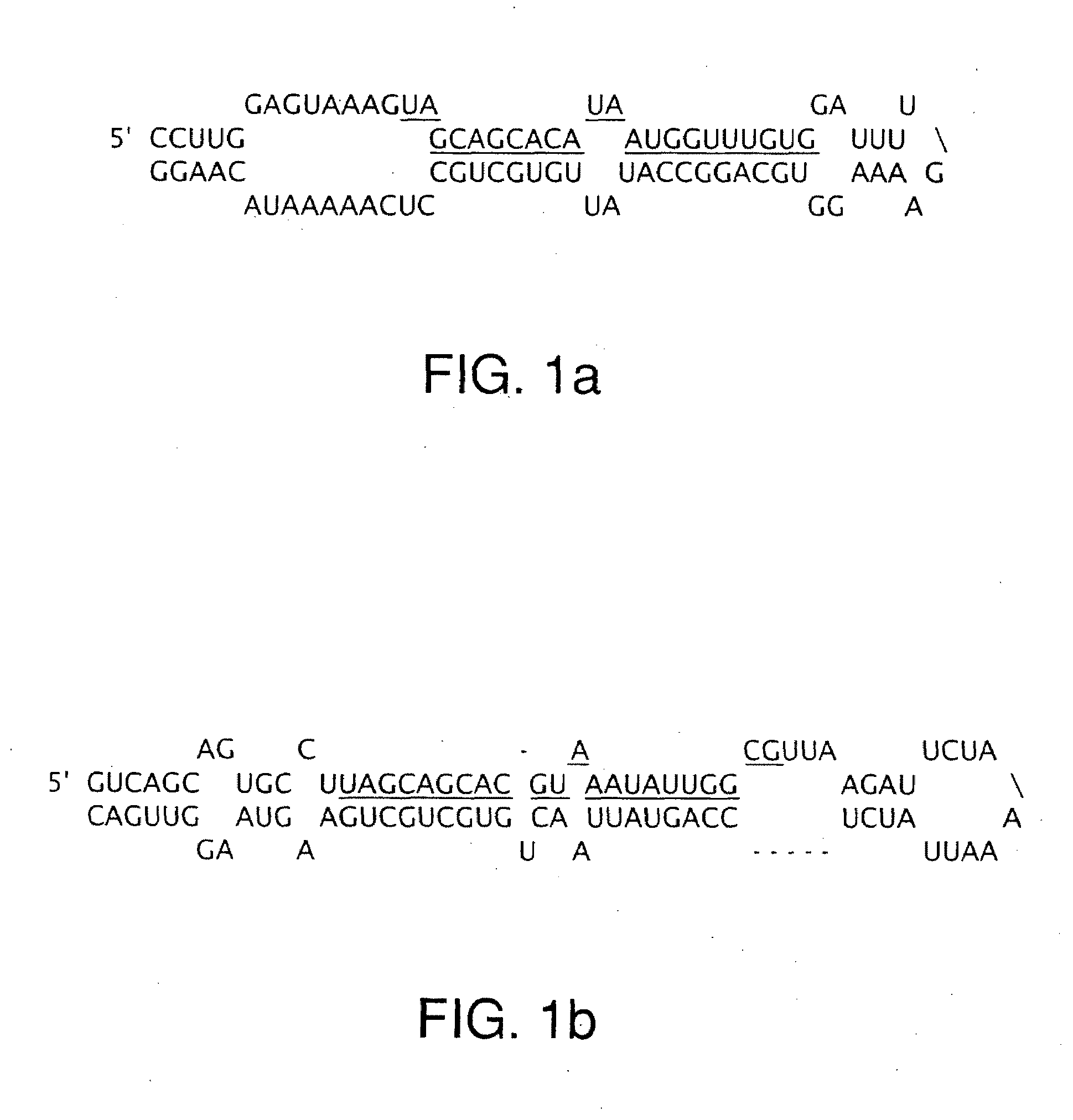
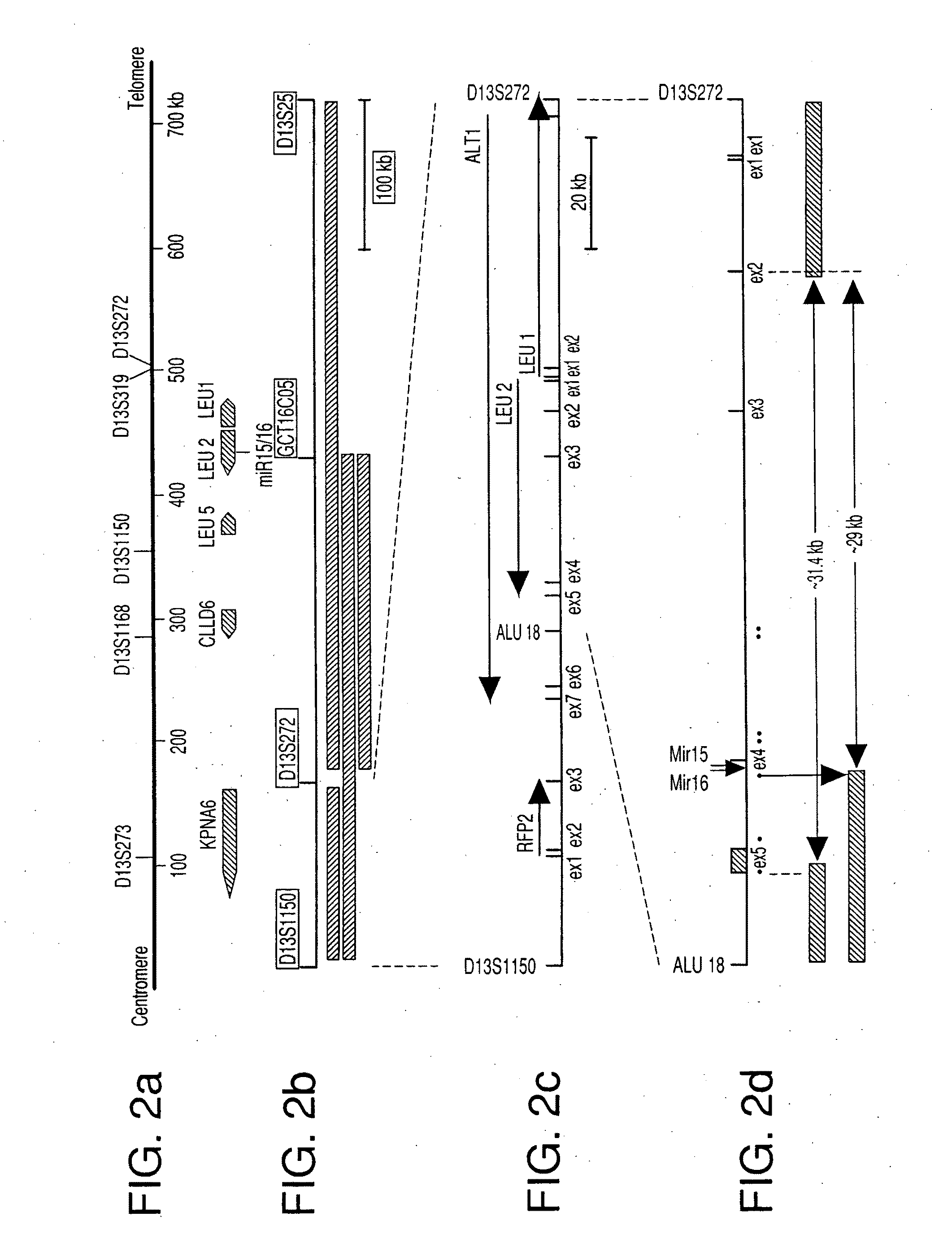
The miR15 and miR16 micro RNA genes are located at 13q14 within a 30 kb region of loss characteristic of cells from certain cancers, such as cells from chronic lymphocytic leukemia or prostate cancer. Chronic lymphocytic leukemia or prostate cancer can be diagnosed by detecting a reduction in miR15 or miR16 gene copy number, by determining miR15 or miR16 gene mutational status, or by detecting a reduction in the RNA transcribed from these genes. The miR15 or miR16 gene products can inhibit the neoplastic or tumorigenic growth of cancers such as chronic lymphocytic leukemia or prostate cancer cells when administered to subjects suffering from these diseases.
Owner:THOMAS JEFFERSON UNIV
Systems, methods and software arrangements for detection of genome copy number variation
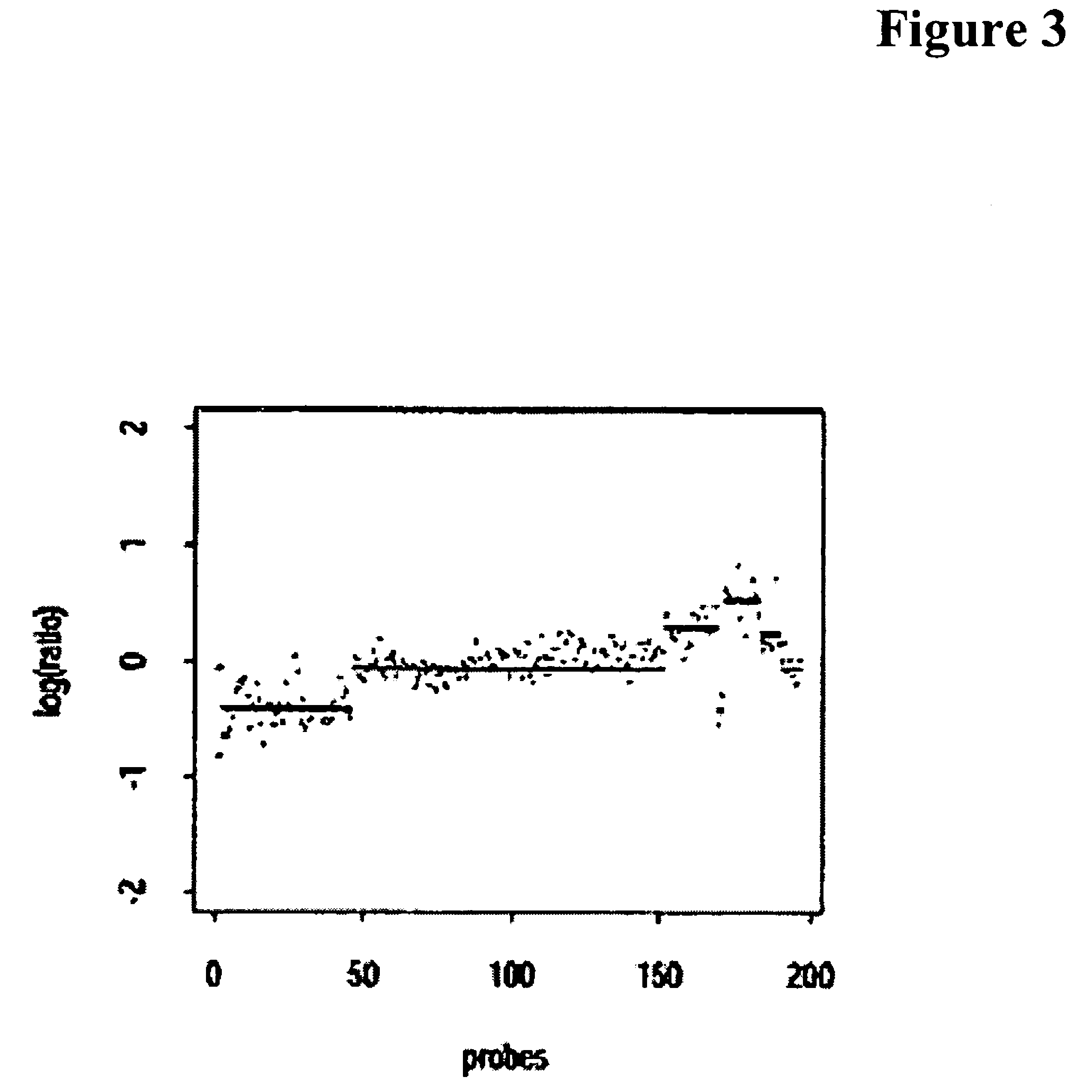
The present invention relates to systems, methods and software arrangements for the detection of variations in the copy number of a gene in a genome. These systems, methods and software arrangements are based on a simple prior model that uses a first process generating amplifications and deletions in the genome, and a second process modifying the signal obtained to account for the corrupting noise inherent in the technical methodology used to scan the genome. A Bayesian approach according to the present invention determines, e.g., the most plausible hypothesis of regional changes in the genome and their associated copy number. The systems, methods, and software arrangements can be are framed as optimization problems, in which a score function is minimized. The system, methods and software arrangements may be useful to assist the scientific study, diagnosis and / or treatment of any disease which has a genetic component, including but not limited to cancers and inherited diseases.
Owner:NEW YORK UNIV
Assays for the direct measurement of gene dosage
InactiveUS20070087345A1Reducing agentMicrobiological testing/measurementIndividual geneImage resolution
The present invention relates to compositions, methods, and kits for quantifying variations in gene copy number, e.g., of individual genes or of chromosomes or portions of chromosomes in an homogeneous reaction, without the need for target amplification, fragment size resolution, or microscopy.
Owner:THIRD WAVE TECH
Methods for Prediction of Clinical Outcome to Epidermal Growth Factor Receptor Inhibitors by Cancer Patients
InactiveUS20080090233A1Raise the possibilityMicrobiological testing/measurementPreparing sample for investigationTyrosineFhit gene
Disclosed are biomarkers, methods and assay kits for the identification of cancer patients who are predicted to benefit, or not to benefit, from the therapeutic administration of an epidermal growth factor receptor (EGFR) inhibitor. The biomarkers of the present invention include detection of EGFR and HER 2 gene amplification and polysomy, EGFR protein expression, EGFR mutations, phosphorylated Akt protein expression, and various combinations of such biomarkers, as well as the combination of these biomarkers with mutations in the tyrosine kinase domain of the EGFR gene. Increased EGFR gene copy number, increased HER2 gene copy number, increased EGFR, protein expression, activated AKT protein expression (phosphorylated AKT) and EGFR mutations are all associated with better outcome for cancer patients treated with EGFR inhibitors. The invention provides a diagnostic paradigm based on each of these tests and combinations of these tests to select cancer patients who will benefit from EGFR inhibitor therapy, as well as a diagnostic paradigm to select cancer patients who will not benefit from EGFR inhibitor therapy.
Owner:UNIV OF COLORADO THE REGENTS OF
Compositions and methods for cancer diagnosis and therapy
The miR15 and miR16 micro RNA genes are located at 13q14 within a 30 kb region of loss characteristic of cells from certain cancers, such as cells from chronic lymphocytic leukemia or prostate cancer. Chronic lymphocytic leukemia or prostate cancer can be diagnosed by detecting a reduction in miR15 or miR16 gene copy number, by determining miR15 or miR16 gene mutational status, or by detecting a reduction in the RNA transcribed from these genes. The miR15 or miR16 gene products can inhibit the neoplastic or tumorigenic growth of cancers such as chronic lymphocytic leukemia or prostate cancer cells when administered to subjects suffering from these diseases.
Owner:THOMAS JEFFERSON UNIV
Transgenic microbial polyhydroxyalkanoate producers
InactiveUS6913911B2Simple production processInduce expressionSugar derivativesBacteriaBiotechnologyTransposon mutagenesis
Transgenic microbial strains are provided which contain the genes required for PHA formation integrated on the chromosome. The strains are advantageous in PHA production processes, because (1) no plasmids need to be maintained, generally obviating the required use of antibiotics or other stabilizing pressures, and (2) no plasmid loss occurs, thereby stabilizing the number of gene copies per cell throughout the fermentation process, resulting in homogeneous PHA product formation throughout the production process. Genes are integrated using standard techniques, preferably transposon mutagenesis. In a preferred embodiment wherein mutiple genes are incorporated, these are incorporated as an operon. Sequences are used to stabilize mRNA, to induce expression as a function of culture conditions (such as phosphate concentration), temperature, and stress, and to aid in selection, through the incorporation of selection markers such as markers conferring antibiotic resistance.
Owner:CJ CHEILJEDANG CORP
Method for predicating homologous recombination deficiency mechanism and method for predicating response of patients to cancer therapy
InactiveCN107287285AInnovativeOvercoming the pitfalls of inaccurate forecastsMicrobiological testing/measurementSequence analysisAbnormal tissue growthPolymerase L
The invention discloses a method for predicating a homologous recombination deficiency (HRD) mechanism and a method for predicating response of patients to cancer therapy and relates to the field of biological information predication. The method comprises the step of judging whether a tumor sample has homologous recombination deficiency or not according to one or more comprehensive values in a large-segment INDEL (Insertion / Deletion) fraction, a copy number variation fraction and a tumor mutation load fraction, wherein the comprehensive values can also comprise a loss of heterozygosity variation fraction. By adopting the method disclosed by the invention, predication of a chromosome large-segment structure, a chromosome gene type number, a chromosome gene copy number, a chromosome variation interval and abnormal loss of heterozygosity and chromosome telomeric imbalance is realized, so that an evaluation range is more complete and HRD can be accurately predicated; the comprehensive values also can be used for determining whether the patients have response to a therapeutic regimen containing one or more of a PARP (Poly Adenosine Diphosphate Ribose Polymerase) inhibitor, an DNA (Deoxyribonucleic Acid) injury inhibitor, a topoisomerase II / II+inhibitor, a topoisomerase I inhibitor and radiotherapy; the method is simple and has wide general applicability.
Owner:SHANGHAI ORIGIMED CO LTD
Transgenic microbial polyhydroxyalkanoate producers
Transgenic microbial strains are provided which contain the genes required for PHA formation integrated on the chromosome. The strains are advantageous in PHA production processes, because (1) no plasmids need to be maintained, generally obviating the required use of antibiotics or other stabilizing pressures, and (2) no plasmid loss occurs, thereby stabilizing the number of gene copies per cell throughout the fermentation process, resulting in homogeneous PHA product formation throughout the production process. Genes are integrated using standard techniques, preferably transposon mutagenesis. In a preferred embodiment wherein mutiple genes are incorporated, these are incorporated as an operon. Sequences are used to stabilize mRNA, to induce expression as a function of culture conditions (such as phosphate concentration), temperature, and stress, and to aid in selection, through the incorporation of selection markers such as markers conferring antibiotic resistance.
Owner:METABOLIX
Genomic classification of colorectal cancer based on patterns of gene copy number alterations
The invention is directed to methods and kits that allow for classification of colorectal cancer cells according to genomic profiles, and methods of diagnosing, predicting clinical outcomes, and stratifying patient populations for clinical testing and treatment using the same.
Owner:ABBVIE INC
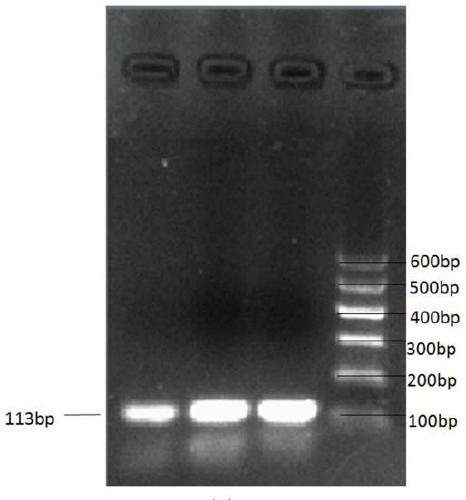
Method for detecting gene copy number variation
ActiveCN102409088ASimple designThe result is accurateMicrobiological testing/measurementPrenatal diagnosisWild type
Owner:郭奇伟 +1
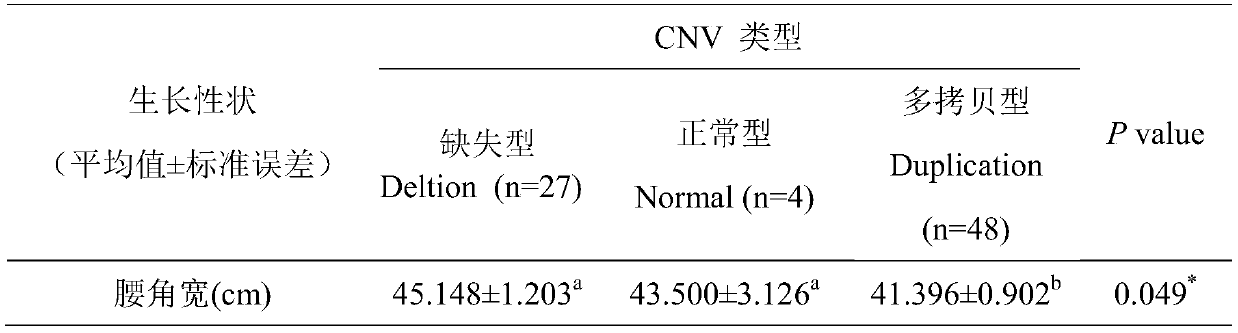
Method for quickly detecting cattle MLLT10 gene CNV marker and application of method
ActiveCN109943647AAccurate identificationLow costMicrobiological testing/measurementMarker-assisted selectionPcr ctpp
The invention discloses a method for quickly detecting a cattle MLLT10 gene CNV marker and application of the method. Based on the real-time fluorescent quantitation PCR technology, cattle genome DNAserves as a template, the cattle MLLT10 gene copy number variation area is amplified by using specific PCR primers, and partial fragments of the cattle BTF3 gene are amplified to serve as an internalreference, and finally, by using the 2-delta delta Ct method, the individual copy number variation type is computed and determined. The method lays a basis for the relationship between the cattle MLLT10 gene copy number variation and the growth trait, the detection method is simple and fast, the method can be used for accelerating cattle marker-assisted selection breeding work, and application andpopularization are facilitated.
Owner:NORTHWEST A & F UNIV
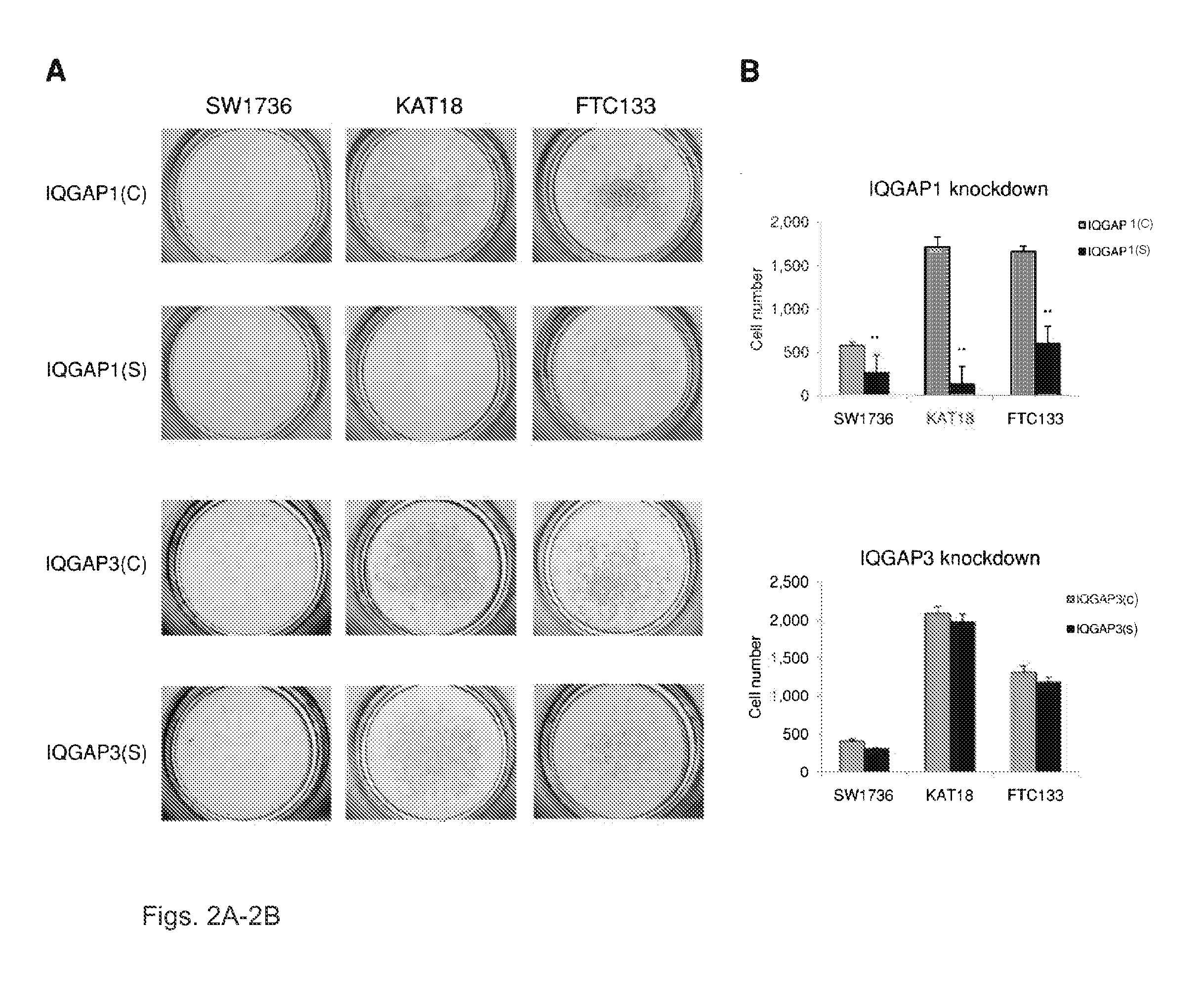
Genetic amplification of IQGAP1 in cancer
ActiveUS9157123B2Diminish invasivenessReduce spreadOrganic active ingredientsSugar derivativesCell invasionFollicular thyroid cancer
We examined IQGAP1 copy gain and its relationship with clinicopathologic outcomes of thyroid cancer and investigated its role in cell invasion and molecules involved in the process. We found IQGAP1 copy number (CN) gain ?3 in 1 of 30 (3%) of benign thyroid tumor, 24 of 74 (32%) follicular variant papillary thyroid cancer (FVPTC), 44 of 107 (41%) follicular thyroid cancer (FTC), 8 of 16 (50%) tall cell papillary thyroid cancer (PTC), and 27 of 41 (66%) anaplastic thyroid cancer, in increasing order of invasiveness of these tumors. A similar tumor distribution trend of CN ?4 was also seen. IQGAP1 copy gain was positively correlated with IQGAP1 protein expression. It was significantly associated with extrathyroidal and vascular invasion of FVPTC and FTC and, remarkably, a 50%-60% rate of multifocality and recurrence of BRAF mutation-positive PTC (P=0.01 and 0.02, respectively). The siRNA knock-down of IQGAP1 dramatically inhibited thyroid cancer cell invasion and colony formation. Co-immunoprecipitation assay showed direct interaction of IQGAP1 with E-cadherin, a known invasion-suppressing molecule, which was upregulated when IQGAP1 was knocked down. IQGAP1, through genetic copy gain, plays an important role in the invasiveness of thyroid cancer and represents a useful prognostic marker and therapeutic target for this and other cancers.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
High-throughput sequencing detection method for gene copy number and gene mutation based on MLPA
InactiveCN105969843AEasy to detectMicrobiological testing/measurementCapillary electrophoresisElectrophoresis
The invention discloses high-throughput sequencing detection technology for gene copy number variation and gene mutation based on an MLPA technical principle, and belongs to the technical field of gene detection. In an MLPA technical link, a PCR amplification link or a link before or after the PCR amplification link is reconstructed to provide a PCR product with a contact sequence capable of entering high-throughput sequencing for sequencing on a machine. Obtained data are analyzed to get a Reads number of amplification of each probe, and the copy number of a template or a gene mutation situation is calculated. According to the technology, fragments with different lengths can be distinguished without capillary electrophoresis, and the products are designed to have almost same lengths, so that the limit that MLPA can only detect more than 50 loci at one time is greatly broken and complex steps for probe preparation are avoided. The contact sequence can be embedded for distinguishing tag sequences for different samples, and therefore it is ensured that detection of the copy number is of high accuracy, and the high-throughput property of detected loci and detected samples is achieved.
Owner:杨永臣
Genomic classification of non-small cell lung carcinoma based on patterns of gene copy number alterations
InactiveUS20100145893A1Digital data processing detailsMicrobiological testing/measurementAbnormal tissue growthTreatment use
The invention is directed to methods and kits that allow for classification of non-small cell lung carcinoma tumors and cell lines according to genomic profiles, and methods of diagnosing, predicting clinical outcomes, and stratifying patient populations for clinical testing and treatment using the same.
Owner:ABBVIE INC
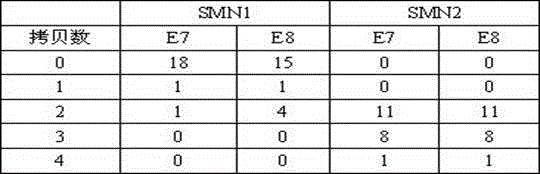
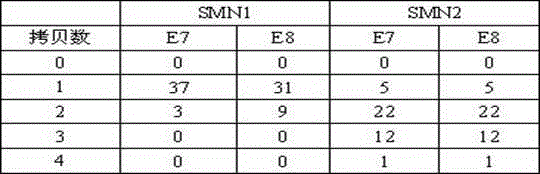
Relative quantitative detection method of human motor neuron gene copy numbers and kit thereof
ActiveCN104630368AIncrease coverageGuaranteed consistencyMicrobiological testing/measurementDNA/RNA fragmentationNucleic acid detectionMedicine
The invention belongs to the technical field of in-vitro nucleic acid detection and especially relates to a relative quantitative detection method of human motor neuron gene copy numbers and a kit thereof. According to the invention, specific primers and probes to the seventh and eighth exons of SMN1 and SMN2 genes and reference gene RPP40 are respectively designed; at the same time, qualitative detection primers and probes are also designed for three hot spot mutations of Y272C, 11bp-DUP and 4bp-DEL of the SMN1 gene. The kit comprises a container containing detection primer and probe compositions, a reference container and a PCR reaction liquid container. Relative quantitative determinations to the copy numbers of the seventh and eighth exons of SMN1 (in the first and second PCR reactions) and the seventh and eighth exons of SMN2 (in the third and fourth PCR reactions) and qualitative detection to the three hot spot mutations of Y272C, 11bp-DUP and 4bp-DEL (in the fifth PCR reactions) are respectively performed by 5 independent PCR reactions. The kit is strong in specificity, high in sensitivity, convenient, fast and applicable to large-scale popularization and application.
Owner:上海春夏正像生物科技有限公司
Systems, methods and software arrangements for detection of genome copy number variation
ActiveUS20060078917A1Weak formEfficient implementationMicrobiological testing/measurementBiostatisticsHypothesisScientific study
The present invention relates to systems, methods and software arrangements for the detection of variations in the copy number of a gene in a genome. These systems, methods and software arrangements are based on a simple prior model that uses a first process generating amplifications and deletions in the genome, and a second process modifying the signal obtained to account for the corrupting noise inherent in the technical methodology used to scan the genome. A Bayesian approach according to the present invention determines, e.g., the most plausible hypothesis of regional changes in the genome and their associated copy number. The systems, methods, and software arrangements can be are framed as optimization problems, in which a score function is minimized. The system, methods and software arrangements may be useful to assist the scientific study, diagnosis and / or treatment of any disease which has a genetic component, including but not limited to cancers and inherited diseases.
Owner:NEW YORK UNIV
Method for gene copy number variation analysis
InactiveCN106055923AHigh-resolutionImprove accuracyHybridisationSpecial data processing applicationsHuman DNA sequencingComputational gene
Owner:WANKANGYUAN TIANJIN GENE TECH CO LTD
Method and system for accurately analyzing DMD gene structural variation breakpoint
ActiveCN107368708AAccurate analysisHigh detection error rateBiostatisticsSequence analysisPositional analysisDmd gene
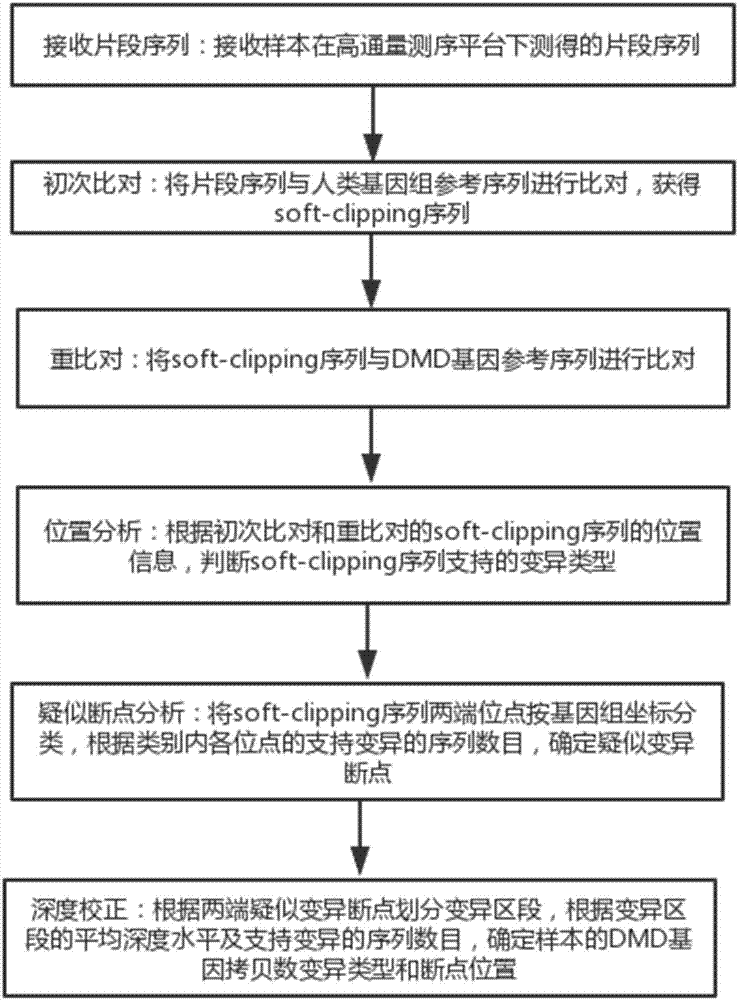
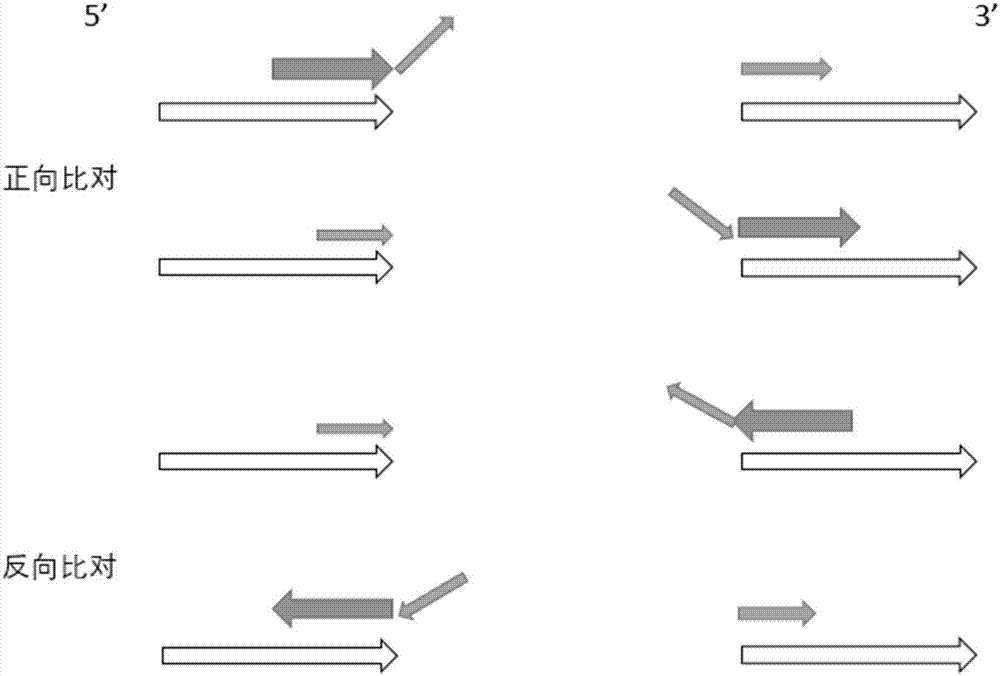
The invention discloses a method and a system for accurately analyzing a DMD gene structural variation breakpoint. The method includes steps of receiving of fragment sequences, initial comparison, repeat comparison, position analysis, analysis of suspected breakpoint, and in-depth correction; through the method, the type and the breakpoint position of DMD gene copy number variation can be accurately analyzed; the mean detection bias rate is less than 4 bp; the accuracy is high, and the stability is good; besides, the method and the system can cover the variation detection of an exon and an introne at the same time, solve the shortcoming that the breakpoint of the gene copy number variation cannot be accurately analyzed in the prior art, and provide technical support for realizing the rapid and parallel DMD breakpoint detection service and the study of the DMD structure gene variation molecular mechanism.
Owner:CAPITALBIO GENOMICS
Multi-copy high expressed recombined plectasin by pichia pastoris
ActiveCN102409003AIncrease expression abundanceFungiAntibody mimetics/scaffoldsPichia pastorisPlectasin
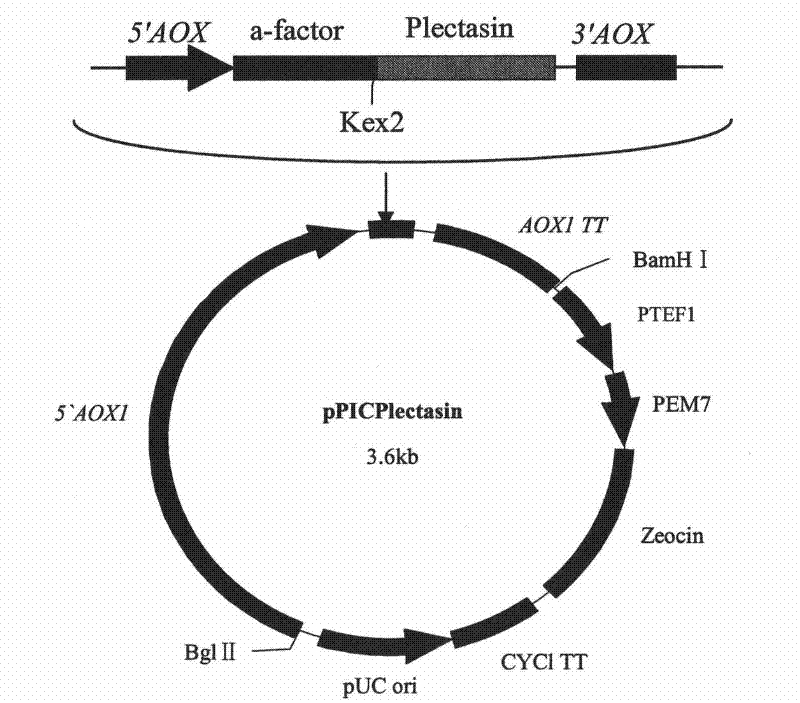
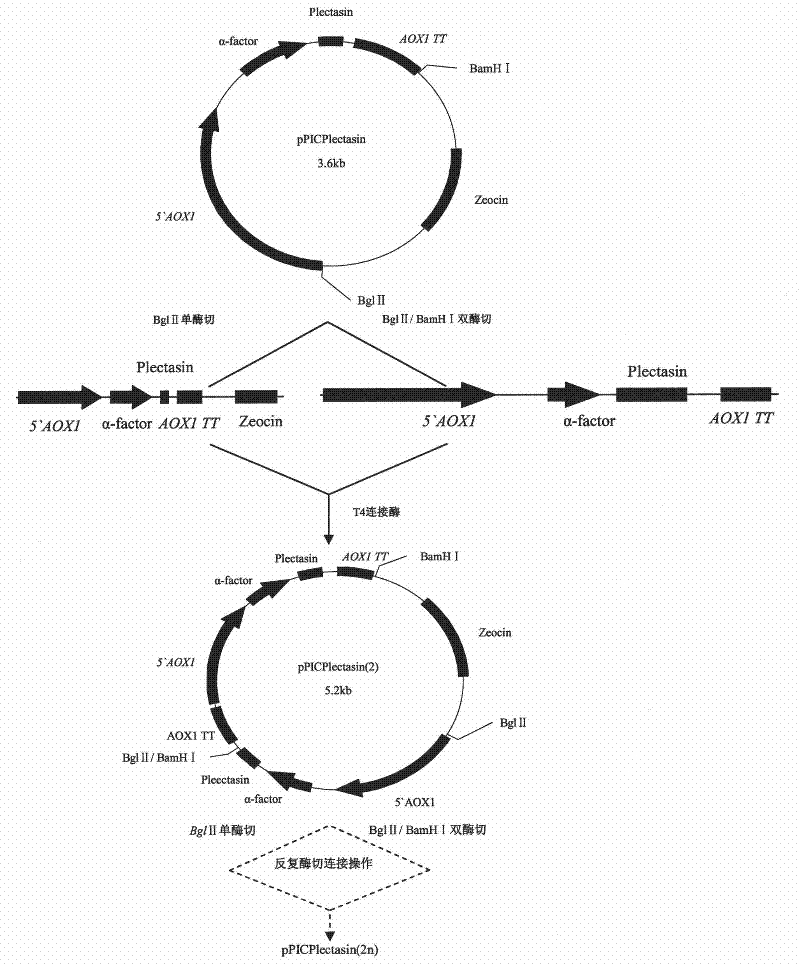
The invention discloses a preparation method of multi-copy high expressed recombined plectasin by pichia pastoris. The method comprises the following steps: a plectasin expressing gene sequence is designed according to preference performance to codon translated by pichia pastoris; the optimized plectasin gene is fused on an alpha-factor signal peptide C terminus of an expression vector pPICZalphaA to construct a single-copy expression vector, the vector comprises a plectasin expression cassette containing a start signal element alcohol oxygen dehydrogenase strong promoter (AOX), alpha-factor signal peptide gene and a plectasin gene fused in C terminus, a stop signal element AOX (TT) and the like. A complementation principle of restriction endonuclease Bg1II and BamHI cohesive end is used to obtain plectasin gene-containing recombinant plasmid of different copy cascade expression cassettes, pichia pastoris is electrotransformed and secreted and expressed plectasin with high efficiency under the methanol induction. The expression level and plectasin gene copy number exist a linear relation. The constructed multi-copy high expressed yeast cells can be used for raising the output and reducing the cost, and is adapted to large scale production of plectasin.
Owner:FEED RESEARCH INSTITUTE CHINESE ACADEMY OF AGRICULTURAL SCIENCES
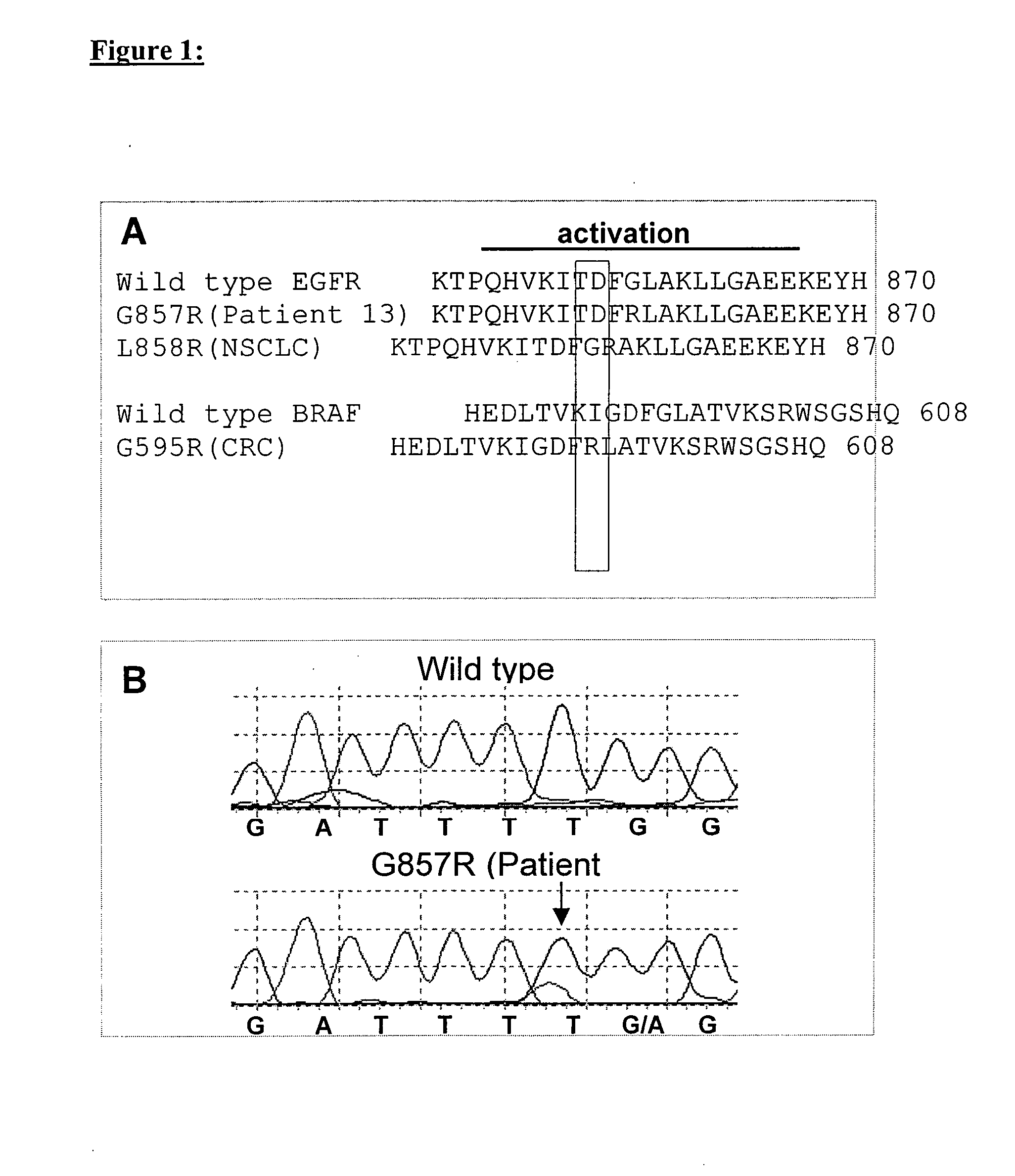
Anti-EGFR antibody therapy based on an increased copy number of the EGFR gene in tumor tissues
InactiveUS20090269344A1Survival predictionHigh gene copy numberMicrobiological testing/measurementAntibody ingredientsAnti-EGFR AntibodyFhit gene
The invention relates to an indidualized and personalized diagnosis and therapy of cancer based on specific molecular alterations which occur in specific tumor tissue of specific tumor patient populations. The therapy and diagnostic is based on the findings that proliferation and tumor growth of specific EGFR bearing tumor tissue expressing an amplified EGFR gene copy number may be abolished by anti-EGFR antibodies, while other individual molecular alterations such as mutations occurring in tumor tissues are unaffected by the same anti-EGFR antibody treatment.
Owner:MERCK PATENT GMBH
Detection method of goat MYLK4 gene CNV marker and application thereof
ActiveCN109355359AThe method for detecting the copy number variation of goat MYLK4 gene is accurate and reliableEasy to operateMicrobiological testing/measurementFluorescenceGenome
The invention discloses a detection method of a goat MYLK4 gene CNV marker and application thereof. The detection method is based on the real-time fluorescence quantification PCR, a goat genome DNA serves as a template, copy number variable regions of goat MYLK4 genes are amplified, then -delta delta Ct calculation is applied, and copy number variation types of individuals are judged according toresults. According to the detection method of a goat MYLK4 gene CNV marker and application thereof, CNV types and growth characteristics of the individuals are used for association analysis; the theoretical basis is laid for establishing the relationship between MYLK4 gene copy number variations and the goat growth characteristics; the detection method is simple, convenient and fast and convenientto apply and popularize, and goat populations with excellent germplasm resources can be quickly established.
Owner:NORTHWEST A & F UNIV
Quantitative detection method for gene copy number
InactiveCN102618646ASync detectionHigh sensitivityMicrobiological testing/measurementReference genesReference sample
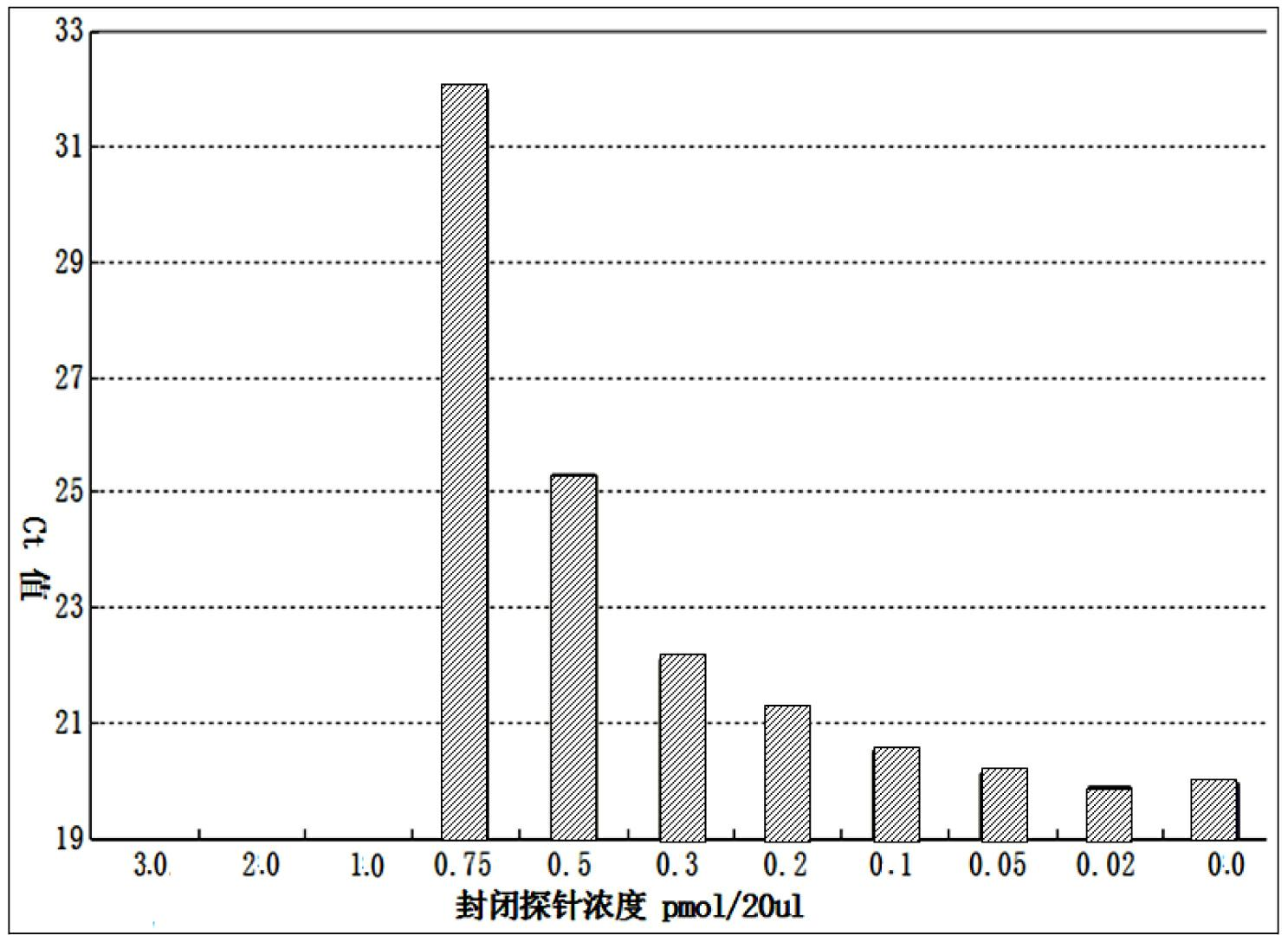
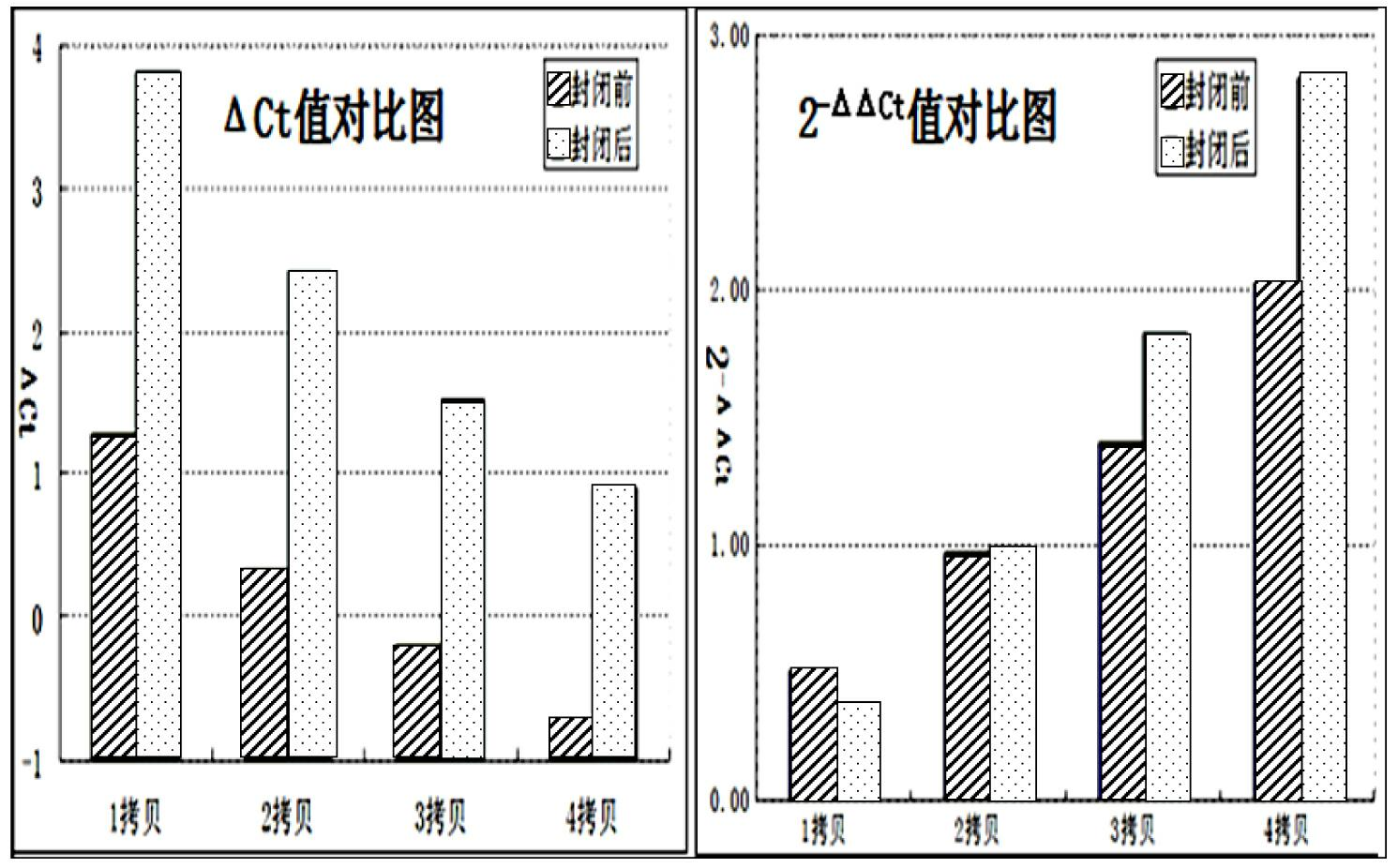
The invention discloses a quantitative detection method for a gene copy number. The method comprises the following steps of: respectively designing primers and fluorescence probes for target gene and reference gene amplification sequences, and designing a closed probe for the target gene amplification sequence; detecting a sample: performing multiple TaqMan fluorescent quantitative polymerase chain reaction (PCR) on target genes and reference genes by using the DNA sample to be detected as a template, and respectively recording Ct values by using a DNA sample with known target gene copy number as a reference sample; and analyzing the result. By the method, the copy number of the functional genes can be directly and quantitatively detected, and deletion, deletion number and multiple copies (CNVs) of the detected genes are directly reflected, so that synchronous quick detection of gene deletion and multiple gene copies is realized; and the method is high in sensitivity, the synchronous quick detection of the gene deletion and the multiple gene copies can be finished only by a common fluorescent quantitative PCR instrument, and experiments prove that the method is accurate and reliable.
Owner:SOUTHERN MEDICAL UNIVERSITY
System and method for detecting copy number variation of cell free tumor genes
ActiveCN108875302AEliminate the effects of identificationEliminate inconsistent capture efficienciesSpecial data processing applicationsCell freeGC-content
The invention provides a system and a method for detecting copy number variation of cell free tumor genes. The average depth of each capture interval is normalized through the average sequencing depthof an overall capture area, so that the influence of change of capture efficiency caused by experiment operation errors on gene copy number identification is eliminated. In a traditional normalization method, a ratio of interval reads to all sequencing reads is adopted as a normalized value of each interval, and the influence on a result caused by inconsistent capture efficiency among different experiments is not eliminated in the traditional method. Through a method for constructing a normal training set, the problem of inconsistent capture efficiency among the capture intervals in samples due to reasons of GC content, probe concentration and the like can be eliminated, so that the false positive rate is reduced.
Owner:广州漫瑞生物信息技术有限公司
Methods for determining the specificity and sensitivity of oligonucleo tides for hybridization
InactiveUS7371516B1High sensitivityStrong specificityBioreactor/fermenter combinationsBiological substance pretreatmentsNucleotideGenomic DNA
The present invention provides materials and methods which may be used to evaluate one or more different probes and select probes that are optimized for sensitivity and specificity for a particular target. In particularly preferred embodiments, the methods and compositions of the invention can be used to evaluate polynucleotide probes having different nucleotide sequences. The methods and compositions thereby allow a user to select a polynucleotide probe, e.g., having a particular nucleotide sequence, that is optimized for sensitivity and / or for specificity for a particular target polynucleotide molecule. In particularly preferred embodiments, the methods and compositions can be used to evaluate a plurality of probes simultaneously, such as on a microarray.Probes evaluated according to the methods of the invention can be selected for and used to detect a variety molecules, including a variety of polynucleotides, such as genomic polynucleotides (e.g., genomic DNA) and genomic transcripts (e.g., mRNAs or cDNA sequences derived from there) as well as gene copy numbers of specific transcripts, single nucleotide polymorphisms, etc.
Owner:MERCK SHARP & DOHME CORP
Method using cattle KCNJ12 gene CNV markers to assist growth trait detection and kit special for method
InactiveCN107523643AThe method for detecting the copy number variation of KCNJ12 gene in cattle is accurate and reliableSimple and fast operationMicrobiological testing/measurementDNA/RNA fragmentationMarker-assisted selectionFluorescence
The invention discloses a method using cattle KCNJ12 gene CNV markers to assist growth trait detection and a kit special for the method. The method is characterized in that the real-time fluorescence quantitative PCR technology is used as the basis, the blood whole genome DNA of Jiaxian red cattle is used as the template, specific primers P1 are used to amplify the copy number variation region of cattle KCNJ12 genes, another pair of specific PCR primers R1 is used to amplify the BTF3 genes of the Jiaxian red cattle to serve as the control group, and the calculation result of 2<-delta delta Ct> is used to judge whether individual copy number has deletion or not. The method is based on the association between the Jiaxian red cattle KCNJ12 gene copy number deletion and growth traits, is beneficial to the acceleration of marker-assisted selection breeding of cattle, and is simple, fast and convenient to popularize and use.
Owner:NORTHWEST A & F UNIV
Method for aided detection of cattle growth characters through copy number variation (CNV) markers on KLF3genes and kit special for aided detection of cattle growth characters through copy number variation (CNV) markers on KLF3genes
InactiveCN107400720AAccurate identificationLow costMicrobiological testing/measurementAnimal scienceZoology
The invention discloses a method for aided detection of cattle growth characters through copy number variation (CNV) markers on a KLF3genes and a kit special for aided detection of the cattle growth characters through the copy number variation (CNV) markers on the KLF3genes. Based on the real-time fluorescent quantitative PCR technology, cattle genome DNA is used as a template, CAN areas of the cattle KLF3genes are amplified, cattle basic transcription factor 3 (BTF3) genes are amplified as the internal reference, and finally the CAN type of individuals is calculated and judged through the 2<-delta delta Ct> method. The method for aided detection of the cattle growth characters through the copy number variation (CNV) markers on the KLF3genes lays a foundation for building the relevance between the KLF3genes CNV and the growth characters of cattle, cattle molecule mark-assist selection breeding work is accelerated advantageously, and the method is simple, rapid and convenient to apply and popularize.
Owner:NORTHWEST A & F UNIV
Construction of saccharomyces cerevisiae engineering bacteria generating beta-phenethyl alcohol, strain and application thereof
InactiveCN103409332ASingle configurationChiral singleFungiMicroorganism based processesAlcohol ethylPhenethyl alcohol
The invention relates to construction of saccharomyces cerevisiae engineering bacteria generating beta-phenethyl alcohol, a strain and application thereof, and belongs to the technical field of bioengineering and production of food spices. The prepared beta-phenethyl alcohol spice is a natural spice and can be used for preparing a plurality of food essences, and therefore, the beta-phenethyl alcohol can be widely applied to industries such as food, medicine and daily use chemicals. Engineering bacterial for coupling overexpression of an aminotransferase gene (ARO8) and decarboxylase gene (ARO10) in the anabolic way of beta-phenethyl are designed and constructed with saccharomycescerevisiae S288 as an original strain; gene copy and expression of the two key enzymes in the main synthetic way are enhanced; the bred engineering bacteria (transformants) are passaged a plurality of times and stable in inheritance; the metabolic flux and the yield of beta-phenethyl are obviously improved. With L-phenylalanine as a major raw material, the engineering bacteria are used for fermenting and converting the raw material into the beta-phenethyl alcohol spice which is single in configuration and chirality and high in fragrance quality, and therefore, a new idea and technical support are provided for preparation of the natural beta-phenethyl alcohol spice.
Owner:BEIJING TECHNOLOGY AND BUSINESS UNIVERSITY
Method for assisting in detecting growth characters by using CNV molecular marker of cattle PLAG1 gene and application thereof
ActiveCN109943646AAccurate identificationLow costMicrobiological testing/measurementClimate change adaptationReference genesMarker-assisted selection
The invention discloses a method for assisting in detecting growth characters by using a CNV molecular marker of cattle PLAG1 genes and an application thereof. The method for assisting in detecting the growth characters by using the CNV molecular marker of the cattle PLAG1 genes comprises the following steps of: respectively amplifying partial fragments of a PLAG1 gene copy number variation regionand a reference gene BTF3 by using a cattle genome DNA as a template through a real-time fluorescence quantitative PCR method, and dividing the quantitative results into multiple copy type and normaltype by 2-delta delta Ct method, thereby identifying copy number variation type of individual PLAG1 gene of a cattle. The copy number variation closely related to the growth characters of the cattleis detected on a DNA level, and the copy number variation can be used as an important candidate molecular marker for marker-assisted selection of the growth characters of Chinese cattles and is used for accelerating the breeding work of the marker-assisted selection of the cattle.
Owner:NORTHWEST A & F UNIV
Method for simply and rapidly identifying transgenic seeds and estimating copy numbers
InactiveCN102229976AEasy to identifyEstimated copy numberMicrobiological testing/measurementFluorescence/phosphorescenceBiotechnologyRapid identification
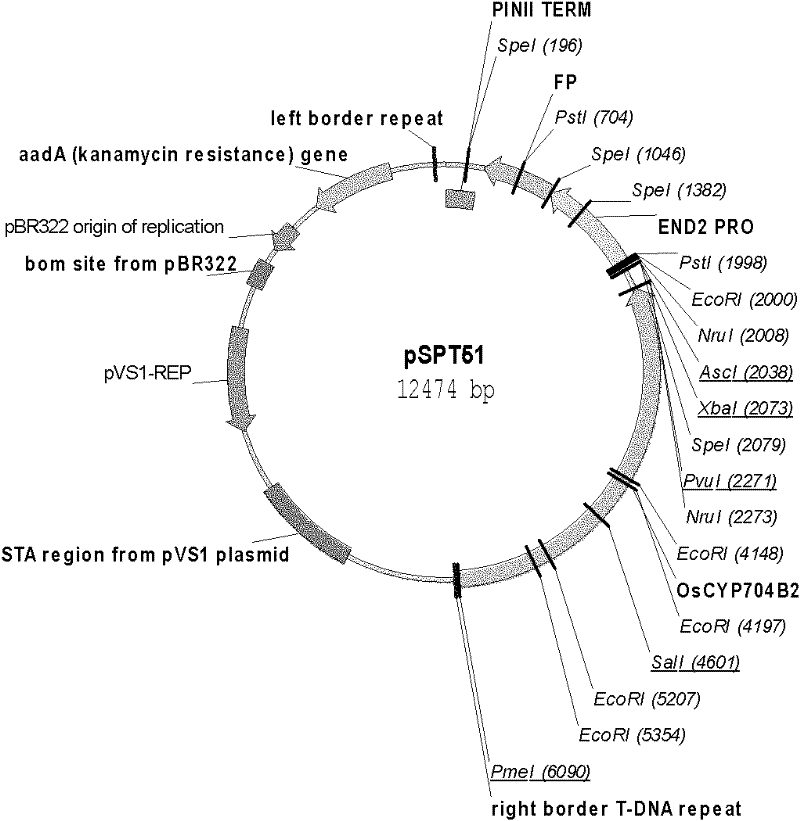
The invention provides a method for simply and rapidly identifying transgenic seeds and estimating copy numbers. According to the invention, the red fluorescent protein (FP) gene (driven by callus / seed coat-specific promoters) is closely connected in series with the target gene and then introduced into the rice male sterile mutant ms26 by means of Agrobacterium tumefaciens-mediated transformation. The transgenic plants are self-crossed to generate the following two types of seeds: non-fluorescent seeds free of transgenic components and fluorescent seeds containing transgenic components. The two types of seeds can be directly separated with the naked eye or with the aid of a fluorescence microscopy, and the separation method is simple and has high accuracy up to 100%. Further, according to the proportions of the fluorescent seeds and the non-fluorescent seeds, the copy numbers of the exogenous gene in the transgenic plant line can be estimated. Since the method provided by the invention does not need the molecular detection of the germinated seeds and young seedlings, the identified seeds can be conveniently applied to the late-stage scientific researches or production.
Owner:BEIJING WEIMING KAITUO CROP DESIGN CENT COMPANYLIMITED +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com