Method for gene copy number variation analysis
A technology for gene copy number and variation analysis, applied in the field of gene information data processing, can solve the problems of high false positive, low accuracy, and no exclusion of copy number variation, so as to reduce false positives, improve accuracy, improve accuracy and The effect of sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0024] It should be noted that, in the case of no conflict, the embodiments of the present invention and the features in the embodiments can be combined with each other.
[0025] The present invention will be described in detail below with reference to the accompanying drawings and examples.
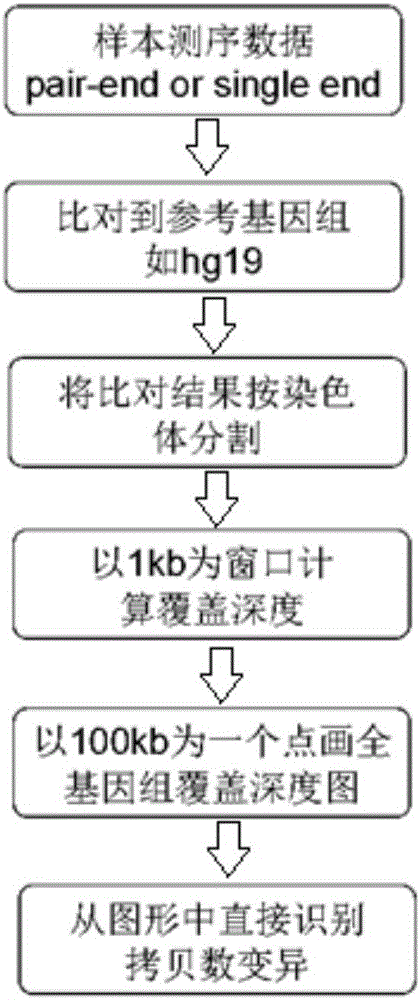
[0026] Such as figure 1 Shown, the implementation steps of the present invention are:
[0027] (1) Read in the index file of the data and the reference genome (two necessary parameters), and complete the analysis of each part in turn.
[0028] (2) The sam file of the comparison result of the entire genome is divided into chromosomes, which can improve the calculation speed and facilitate statistical analysis.
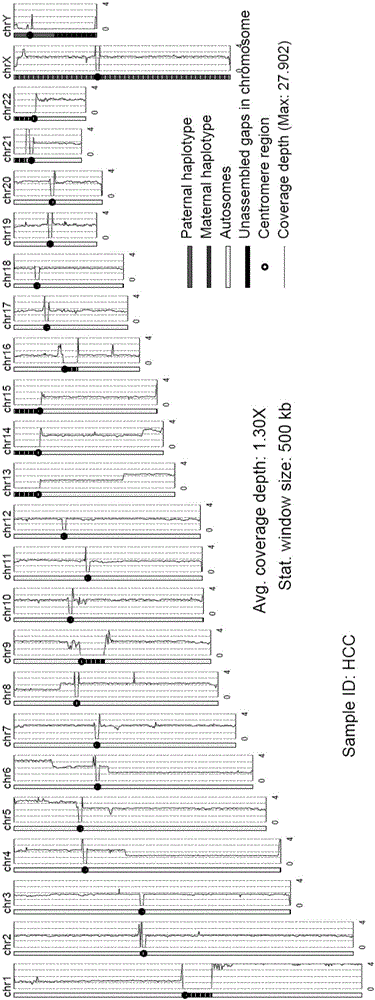
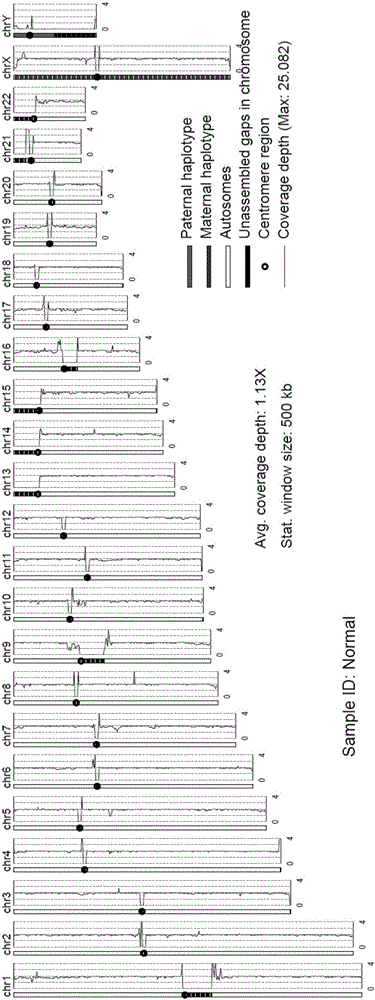
[0029] (3) Comparing and comparing the comparison results of the sequencing data to make statistics. Including the amount of raw data, mapping rate, unique mapping rate, genome coverage, average coverage depth, average length of inserted sequences during library construction, a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com