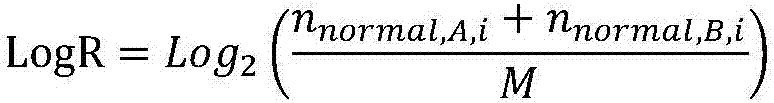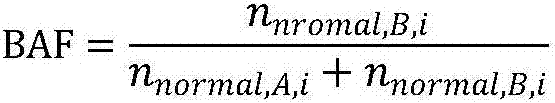Method for predicating homologous recombination deficiency mechanism and method for predicating response of patients to cancer therapy
A technology of homologous recombination and mechanism, applied in the field of biological information, can solve the problems of fixed computing type, lack of homologous recombination mechanism, copy number variation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0174] Example 1 Obtaining the Gene Sequences of Tumor Samples and Normal Samples
[0175] 1. Acquisition of targeted gene bank
[0176] 1.1 Sample processing
[0177] Obtain tumor samples and normal samples from the same patient, perform DNA extraction, and use quantitative instruments (such as Nanodrop instruments) for quantification, so as to determine whether the DNA quality of normal sample DNA and tumor samples meets the subsequent sequencing requirements. The kit is used to extract sample DNA, followed by DNA amplification to obtain sufficient tumor DNA samples and normal DNA samples respectively.
[0178] Wherein, the sample type includes but is not limited to one of FFPE (paraffin-embedded sample), tissue (tissue cut from the patient's body) sample, and blood (blood drawn from the patient's body) sample. According to different sample types, select Different kits are used to extract DNA from normal samples and tumor samples. For example, FFPE samples are extracted wi...
Embodiment 2
[0187] Embodiment 2 Calculation of large fragment INDEL score value
[0188] Calculation of large fragment INDEL score (ie OM-INDEL score): OM-INDEL score is the number of regions where large fragment INDEL (insertion length ≥ 25bp and deletion fragment length ≥ 50bp) occurs.
[0189] The algorithm operation flow is as follows:
[0190] 1. Select high-quality sequencing sequences
[0191] The target gene sequence of the tumor sample obtained in Example 1 and the target gene sequence of the normal sample are respectively subjected to base quality screening, and if a sequencing sequence has one or more base quality values lower than 20, then filter the sequence to obtain the base quality Both tumor sample sequences and normal sample sequences are greater than 20.
[0192] The base quality screening of the present invention is suitable for higher sequencing depths, such as sequences with a sequencing depth greater than 500X-1000X, and can reduce the impact of system sequencin...
Embodiment 3
[0202] Example 3 Calculation of Copy Number Variation Score Values
[0203] The fractional copy number variation value is the number of regions where copy number amplification and copy number loss occurred, ie copy number variation HRD-CNV.
[0204] Using the normal sample comparison result obtained in step 3 of Example 1 and the tumor sample comparison result as input, use copy number variation software to perform CNV (Copy Number Variants, i.e. copy number variation) analysis (for example, EXCAVATOR) to determine the tumor In which regions of the chromosome are amplification and deletion occurred in the sample, the number of amplification and deletion regions is counted, and this number is the HRD-CNV score. When the HRD-CNV score is 0, it means that the sample has no copy number amplification or copy Conversely, when the HRD-CNV score is not 0, it means that the sample has copy number amplification or copy number deletion.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com