Method for quickly detecting cattle MLLT10 gene CNV marker and application of method
A cattle and gene technology, applied in the field of molecular genetics, can solve unseen problems, achieve the effects of low cost, easy popularization and application, and speed up the molecular marker-assisted selection breeding of cattle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0028] The present invention will be described in detail below in conjunction with the accompanying drawings and embodiments, which are only explanations of the present invention, rather than limiting the protection scope of the present invention.
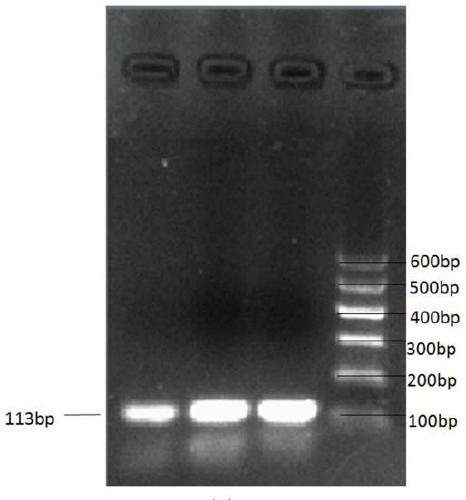
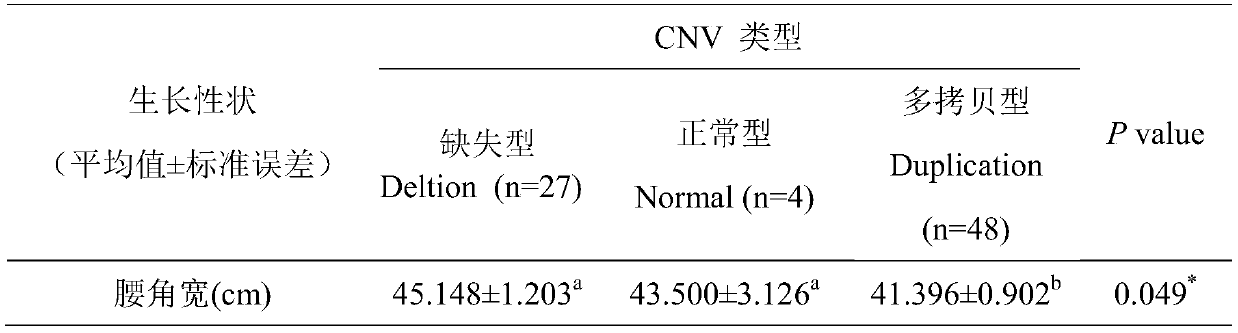
[0029] In the previous cattle genome resequencing study, a copy number variation located in the Chr13:23206001-23210800 region of the non-coding region of the bovine MLLT10 gene was found. Therefore, the present invention designs specific primers based on the region of copy number variation in the cattle MLLT10 gene sequence obtained by resequencing, and then uses the genomic DNA of cattle (Qinchuan cattle, Xianan cattle, Jiaxian cattle, Yunling cattle) as templates, qPCR amplification was carried out, and the BTF3 gene was used as an internal reference gene, using 2 -△△Ct A method for determining the type of copy number variation of an individual. Based on the physiological function of MLLT10 gene and the regulation mechanism of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com