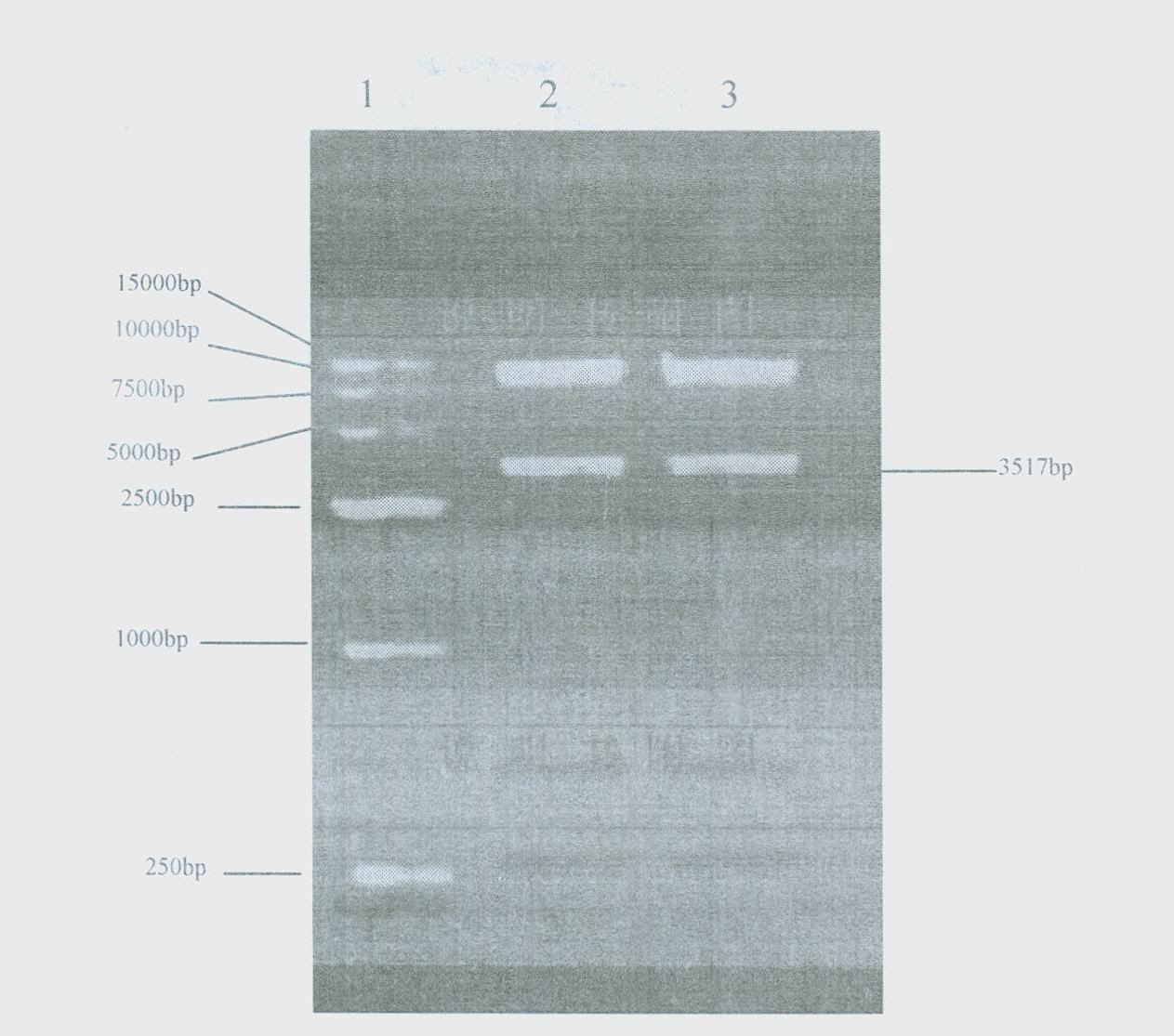
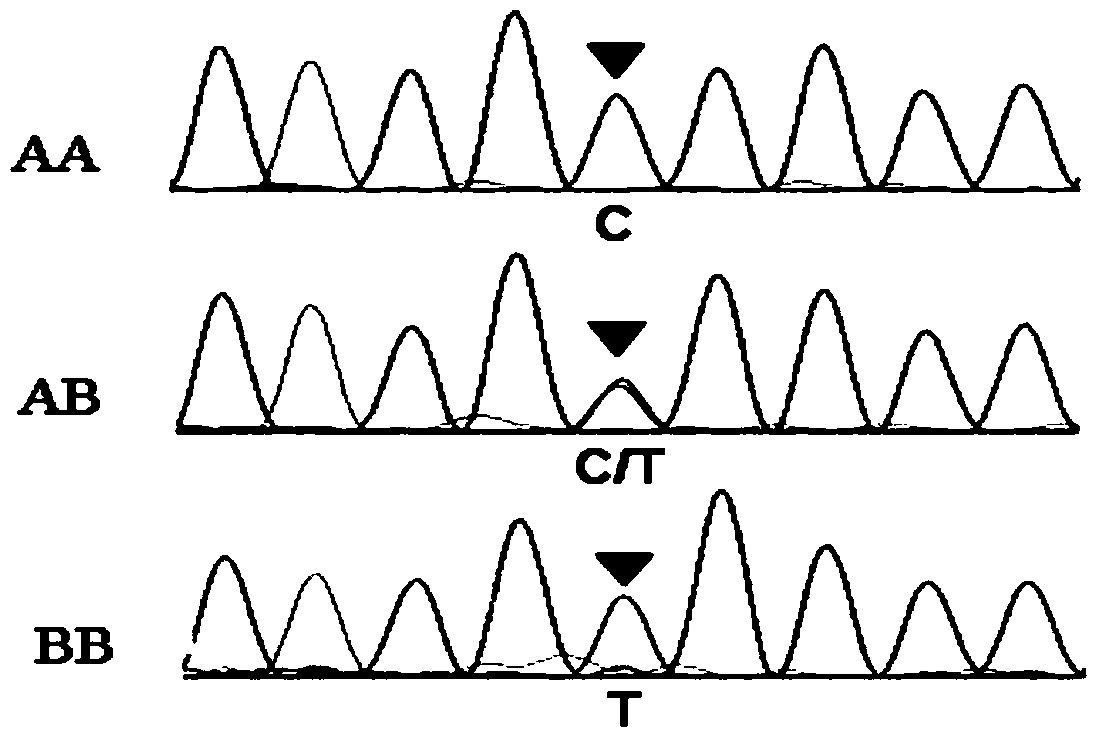
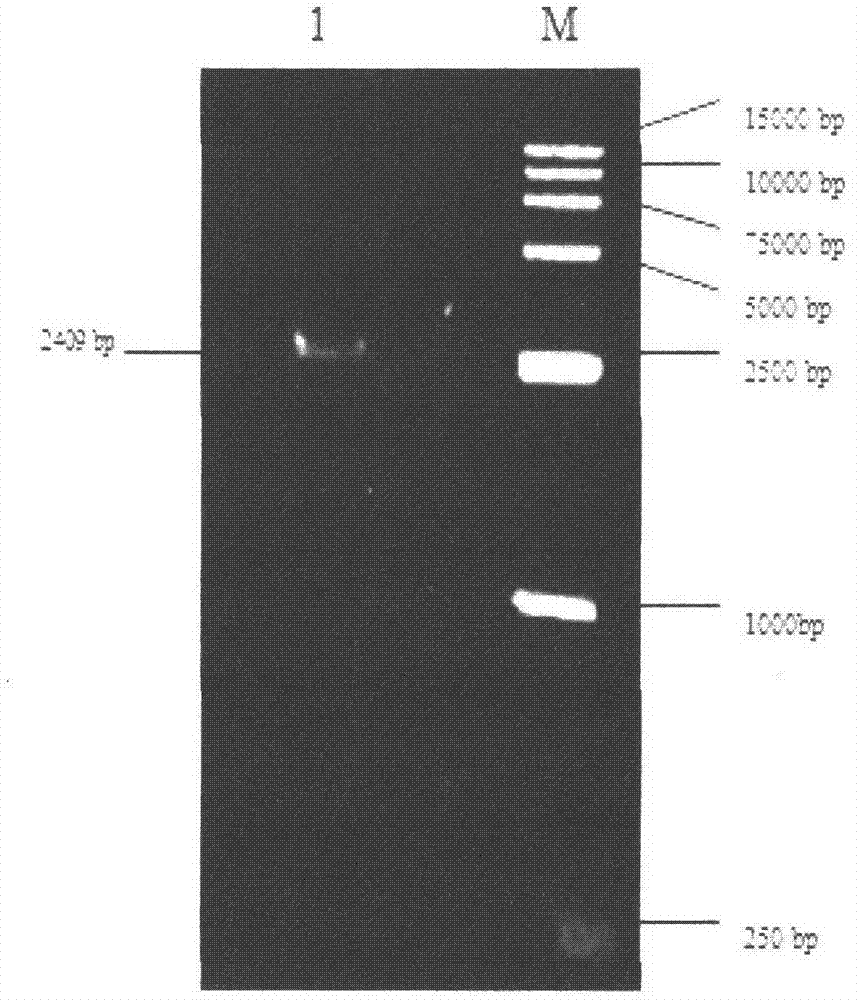
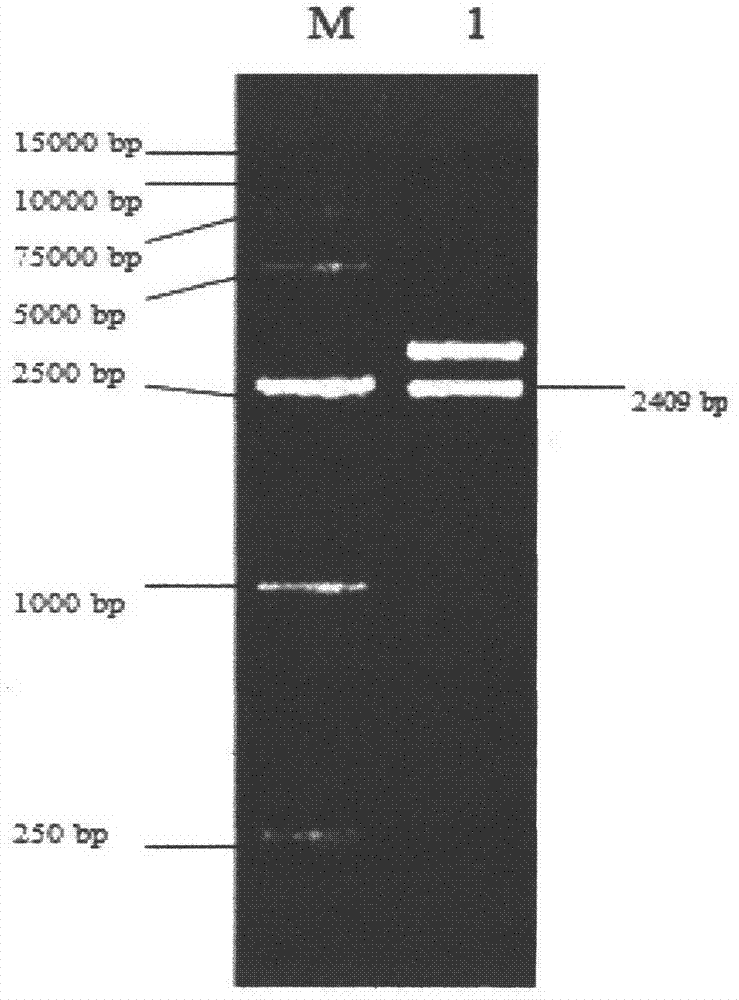
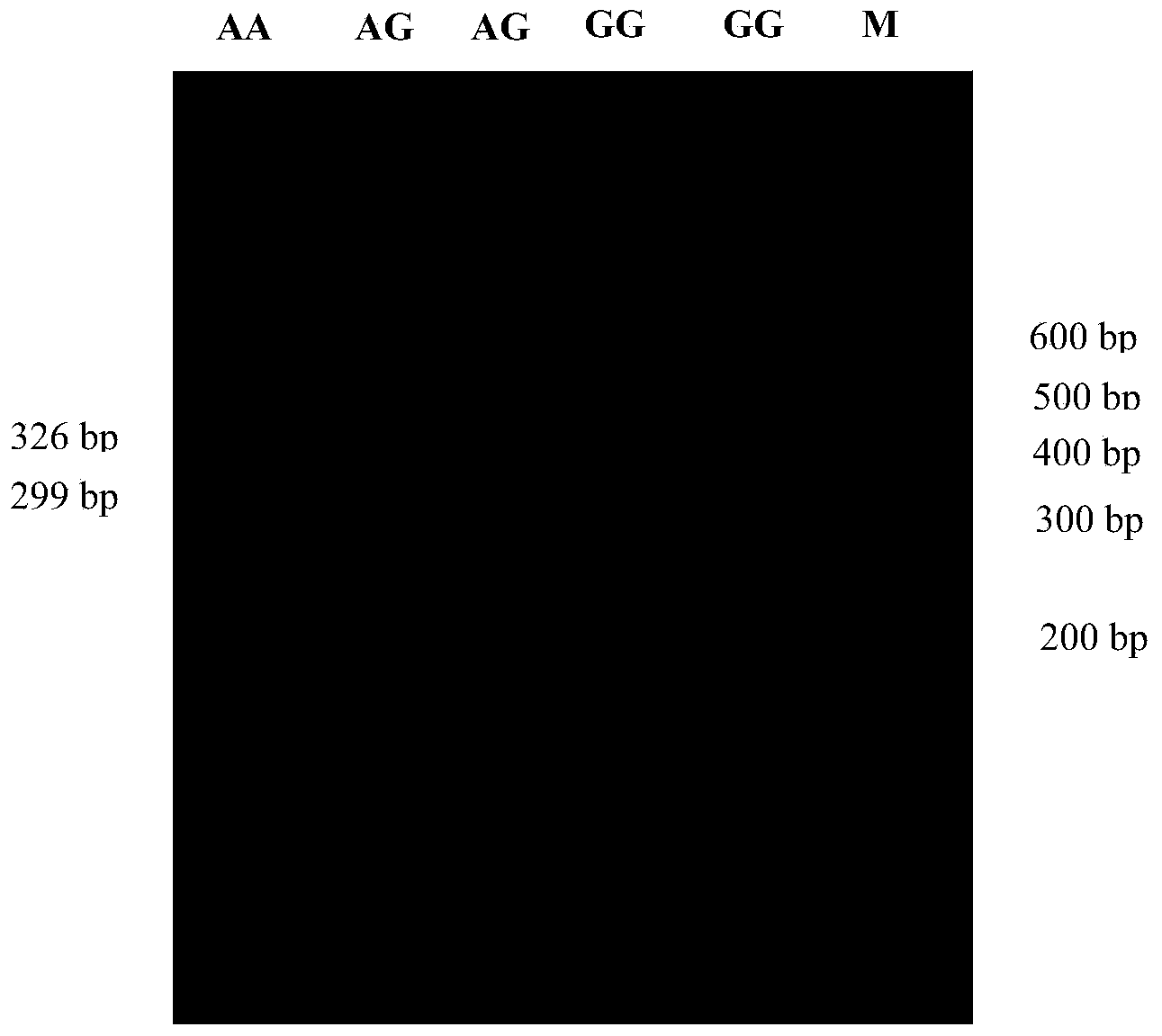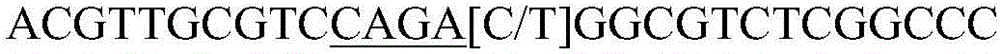Patents
Literature
48 results about "Qinchuan cattle" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
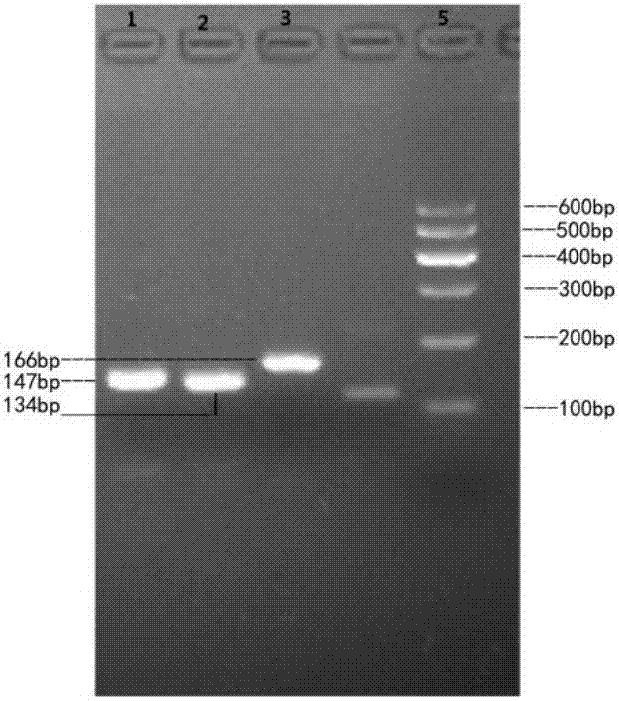
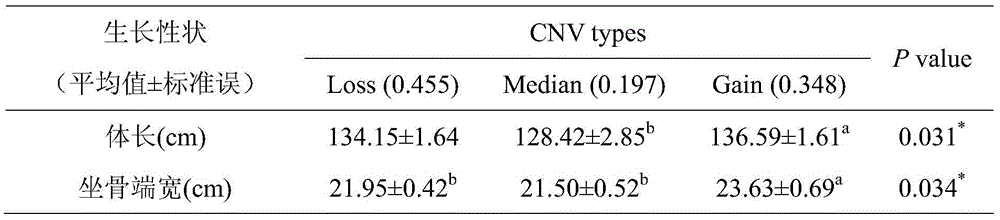
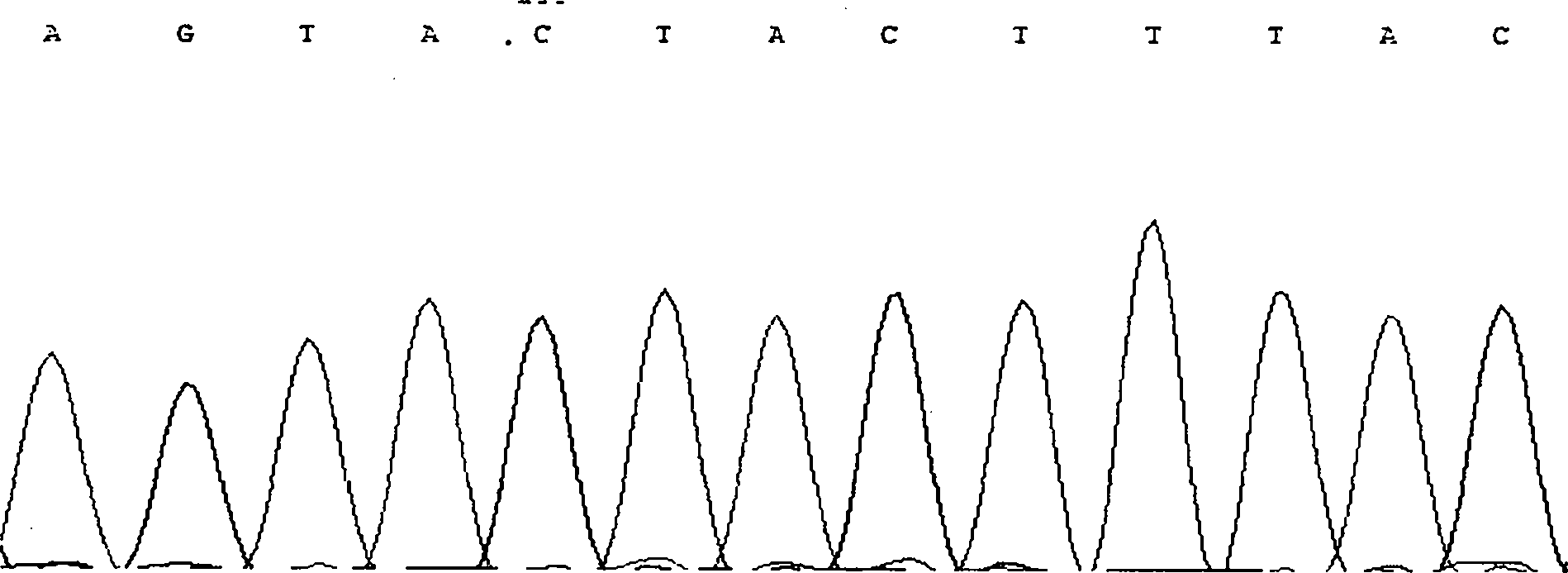
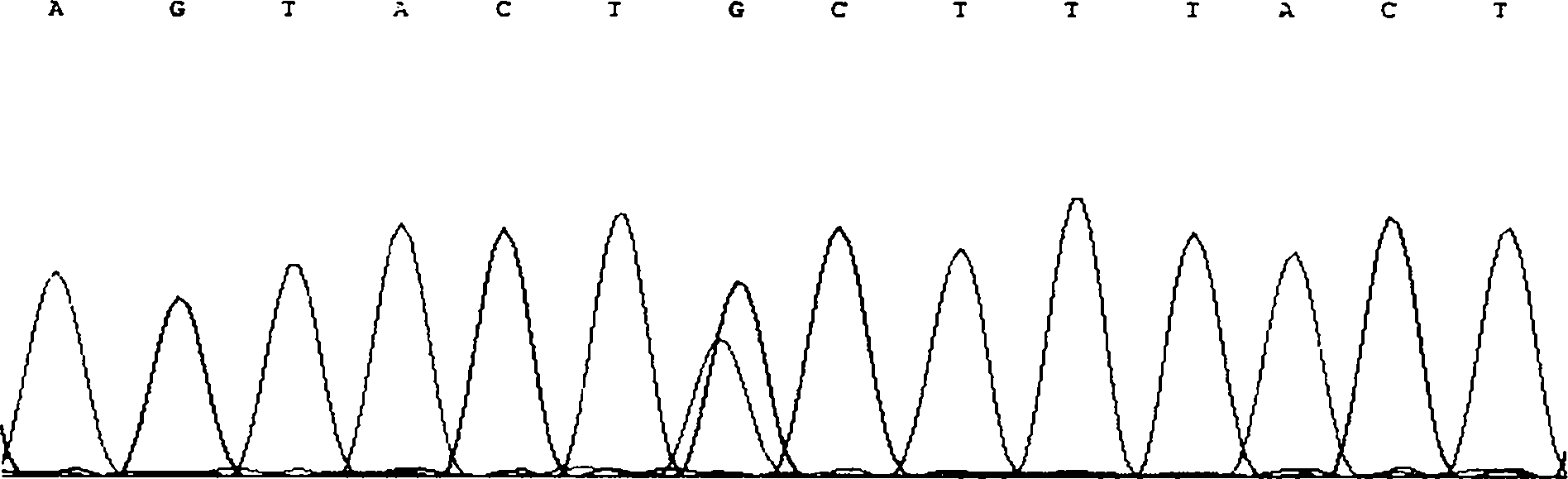
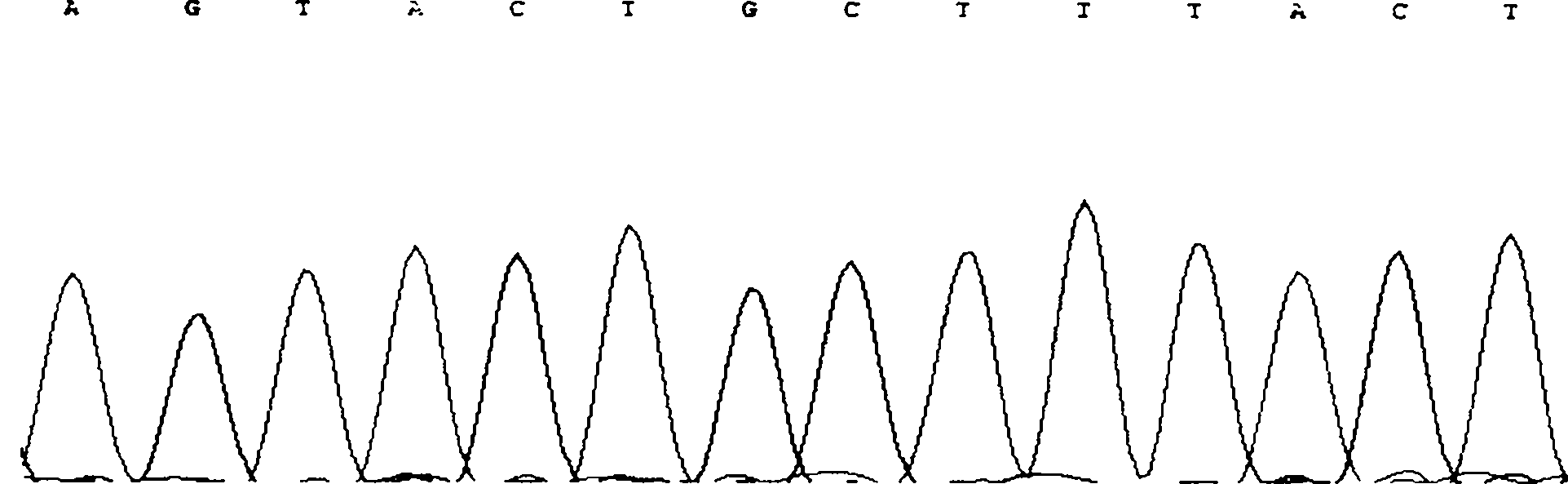
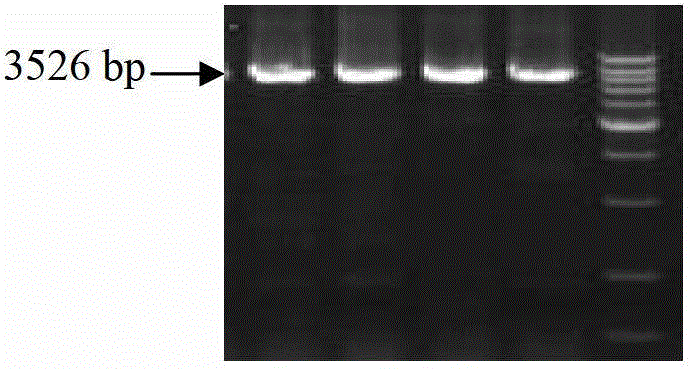
Method for detecting CNV (Copy Number Variation) mark of a gene GBP2 of Qinchuan cattle and application of CNV mark
InactiveCN107119117AThe method for detecting the copy number variation of bovine GBP2 gene is accurate and reliableSimple and fast operationMicrobiological testing/measurementMarker-assisted selectionPcr ctpp
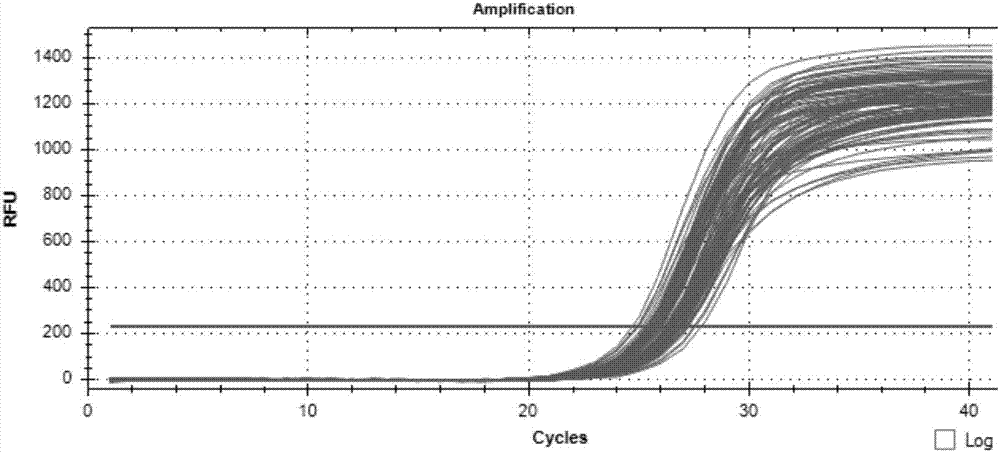
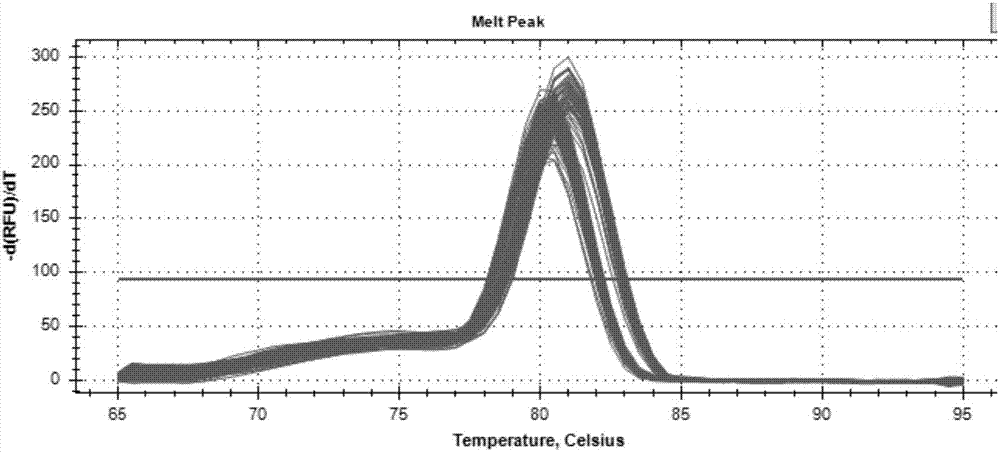
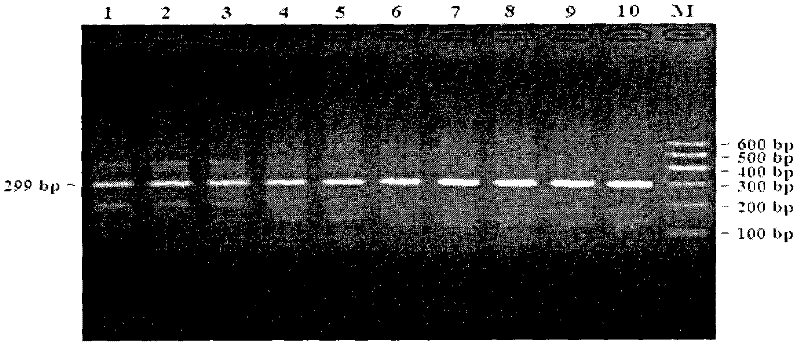
The invention discloses a method for detecting a CNV (Copy Number Variation) mark of a gene GBP2 of Qinchuan cattle and the application of the CNV mark. The method comprises the following steps: on the basis of a real-time fluorescent quantitation PCR (Polymerase Chain Reaction) technology, by taking the DNA of a whole genome of blood of the Qinchuan cattle as a template, respectively multiplying two CNV regions of the gene GBP2 of the cattle by using two pairs of specific primers GBP2-CNV1 and GBP2-CNV2, multiplying a gene BTF3, which serves as a contrast, of the Qinchuan cattle by using another pair of specific PCR primers R1, and judging whether the copy number of individuals is missing or not according to a calculation result of 2-delta delta Ct. According to the method provided by the invention, on the basis of the relevance between missing of the copy number of the gene GBP2 of the Qinchuan cattle and a growth character, the Qinchuan cattle molecular mark assistant selection breeding work is accelerated favorably. The method is simple, quick and convenient for popularization and application.
Owner:NORTHWEST A & F UNIV
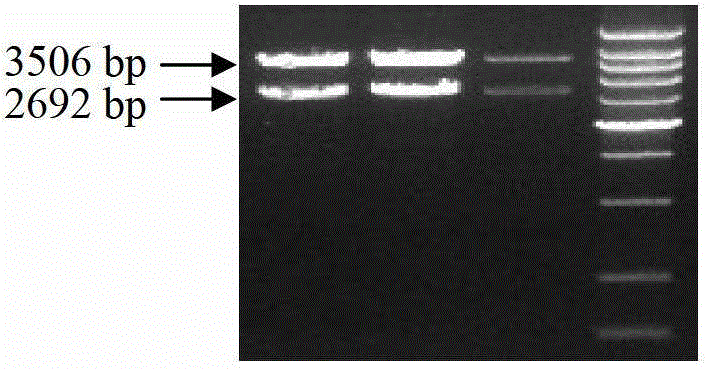
Method of detecting copy number variation of Qinchuan cattle FGF13 genes and application thereof
ActiveCN105543352AAccurate identificationLow costMicrobiological testing/measurementMarker-assisted selectionDNA
The invention discloses a method of detecting copy number variation of Qinchuan cattle FGF13 genes and application thereof. According to the method, based on real-time quantitative PCR, Qinchuan cattle genome DNA is used as a template, a pair of specific PCR primers is utilized for amplifying a copy number variation region of the Qinchuan cattle FGF13 genes, another pair of specific PCR primers is utilized for amplifying cattle general transcription factor 3 genes as reference at the same time, and lastly a 2-delta delta Ct method is utilized for calculating and determining the individual copy number variation type. The method lays a foundation for establishment of correlation between copy number variation of the Qinchuan cattle FGF13 genes and growth traits, and is beneficial to accelerating Qinchuan cattle molecular marker assisted selective breeding work, simple, quick and convenient to apply and popularize.
Owner:NORTHWEST A & F UNIV
Qinchuan cattle FoxO1 gene mononucleotide polymorphism molecular marker detection method and application
InactiveCN103233001AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationNucleotideElectrophoresis
The invention discloses cattle FoxO1 gene mononucleotide polymorphism and a detection method thereof. The gene mononucleotide polymorphism comprises that: with a Qinchuan cattle whole genome DNA comprising the FoxO1 gene as a template, and with a primer pair P as primers, the Qinchuan cattle FoxO1 gene is subjected to PCR amplification; a restriction endonuclease HhaI is used for digesting the PCR amplification product, and the digested segment is subjected to agarose gel electrophoresis; and according to the electrophoresis result, base polymorphism on the 178132 site of the Qinchuan cattle FoxO1 gene is identified. The FoxO1 gene is important for animal muscle fat traits and growth traits. With the method, the molecular genetic marker closely related to Qinchuan cattle growth traits is screened and detected on a DNA level. The method is used in Qinchuan cattle assisted selection and molecular breeding, and assists in accelerating Qinchuan cattle breeding speed.
Owner:NORTHWEST A & F UNIV
Method of applying adenovirus vector mediation ribose nucleic acid (RNA) jamming technology to silence sterol regulatory element binding protein 1
ActiveCN103146752AAccurate measurementUnaffected by subjective judgmentViruses/bacteriophagesFermentationSterolSterol Regulatory Element Binding Protein 1
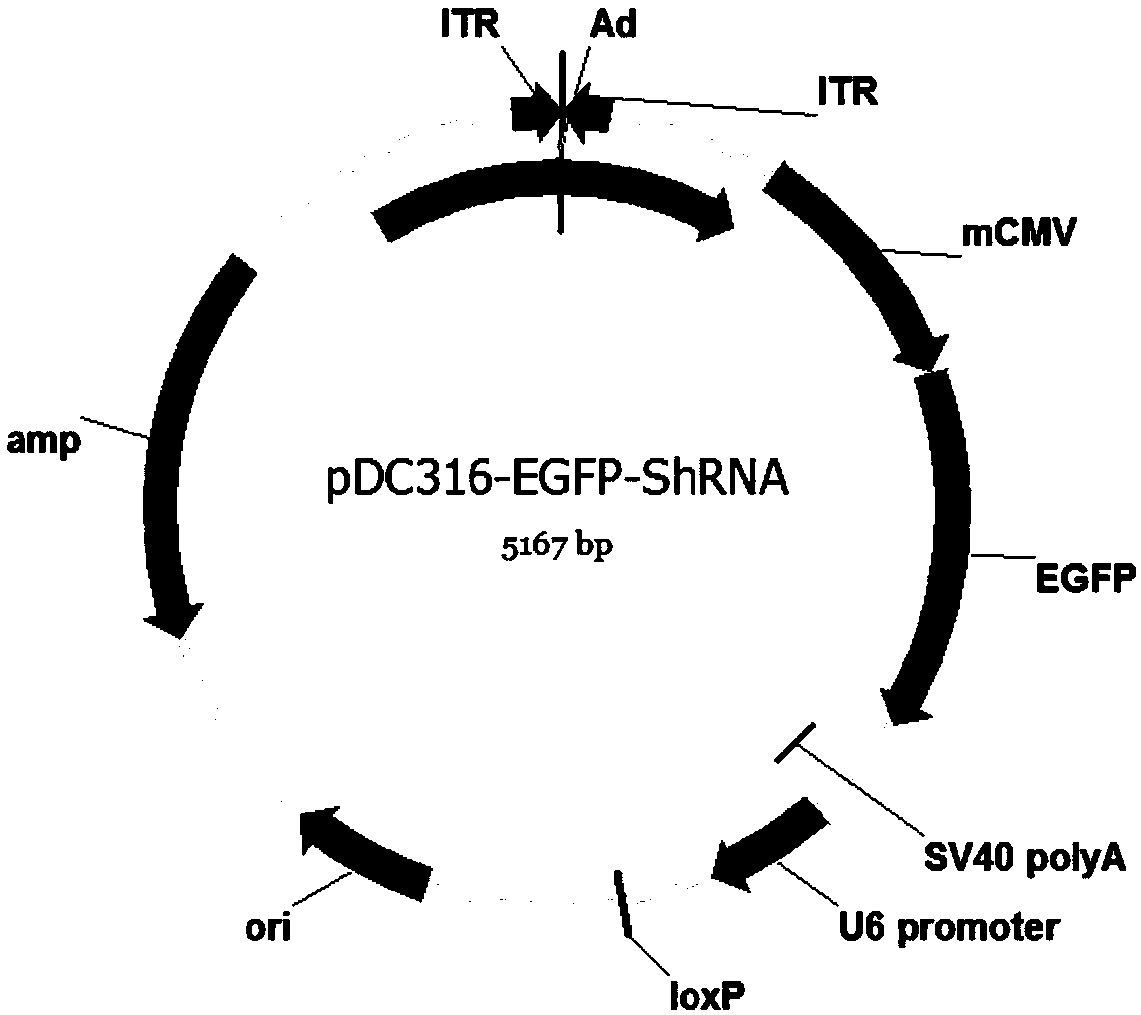
The invention discloses a method of applying an adenovirus vector mediation ribose nucleic acid (RNA) jamming technology to silence a sterol regulatory element binding protein 1. The method of applying the adenovirus vector mediation RNA jamming technology to silence the sterol regulatory element binding protein 1 comprises the following steps: A1, cloning a Qinchuan cattle sterol regulatory element binding protein 1 (SREBP1); A2, constructing a psiCHECK-II-SREBP1 expression vector; A3, designing and synthesizing a target SREBP1 gene sh RNA; A4, constructing a pENTR / CMV-GFP / U6-sh RNA expression vector; A5, determining and screening interference efficiency of sh RNA; A6, constructing a recombinant adenovirous vector; and A7, carrying out adenovirus packaging, amplification and titre determination. The method of applying the adenovirus vector mediation RNA jamming technology to silence the sterol regulatory element binding protein 1 is an effective method of applying the RNA jamming technology to research functions of the Qinchuan cattle SREBP1 gene.
Owner:NORTHWEST A & F UNIV
Method for detecting Qinchuan cattle Wnt7a gene single nucleotide polymorphism, and application thereof
InactiveCN103320429AAccurate detectionLow costMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionMarker-assisted selection
The invention discloses a method for detecting Qinchuan cattle Wnt7a gene single nucleotide polymorphism (SNP). According to the invention, genome DNA of a Chinese local cattle breed Qinchuan cattle is adopted as a template; Wnt7a gene sequence is subjected to PCR amplification by using Taq DNA polymerase, Buffer (a buffer solution), Mg<2+>, dNTPs and primers M; a PCR product is subjected to enzyme digestion by using restriction endonuclease Alw44I; a fragment is subjected to separation and detection through agarose gel electrophoresis, and Qinchuan cattle Wnt7a gene single nucleotide polymorphism is finally determined. The Wnt7a gene function is related to growth traits such as weight and body length, such that the detection method provided by the invention establishes a basis for establishing a relationship between the SNP of the Wnt7a gene and growth traits. Therefore, marker-assisted selection of meat-purpose growth traits of Chinese Qinchuan cattle is facilitated, and a Qinchuan cattle population with excellent genetic resource can be rapidly established.
Owner:NORTHWEST A & F UNIV
Detection method for 19-bp duplication and deletion polymorphism of Chinese yellow cattle PLAG1 (Pleomorphic Adenoma Gene 1) gene and application of detection method
InactiveCN108866202AEasy to detectLow costMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceNanyang cattle
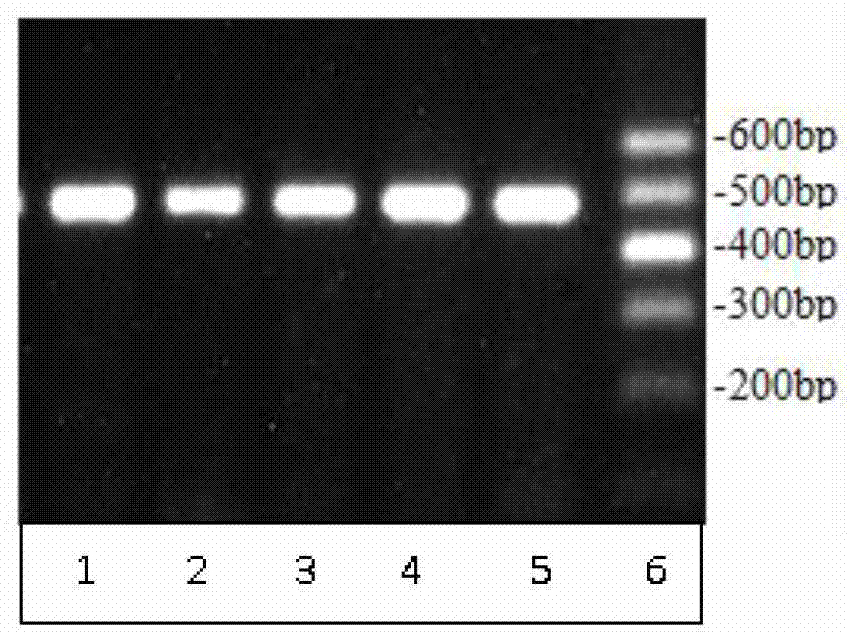
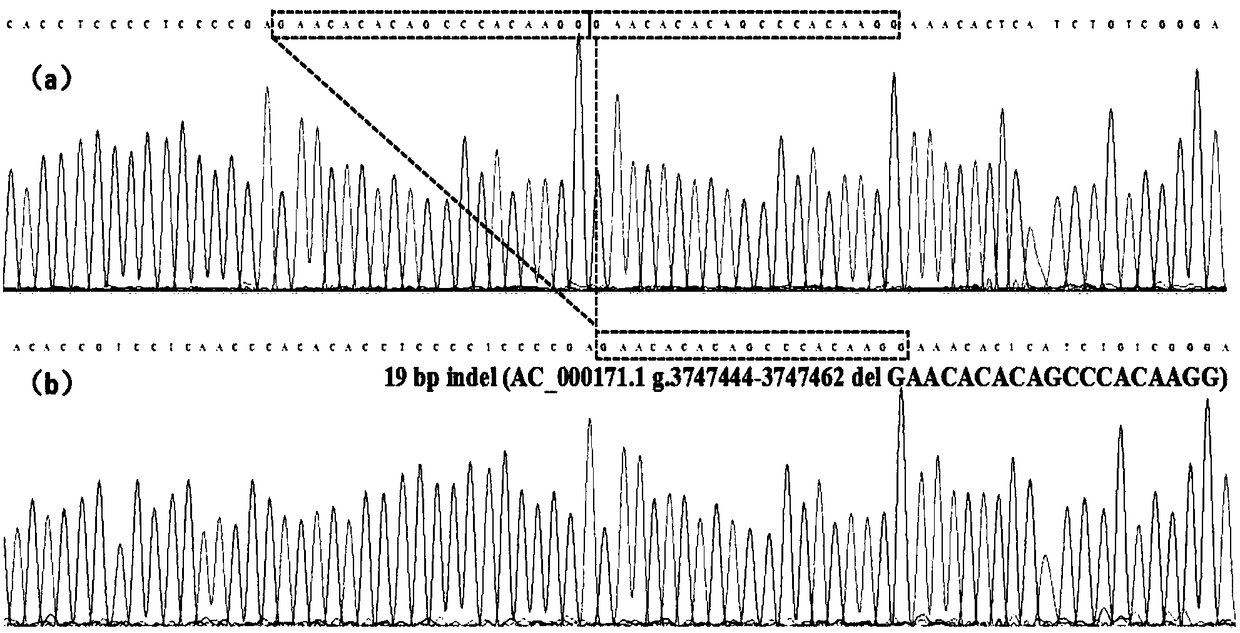
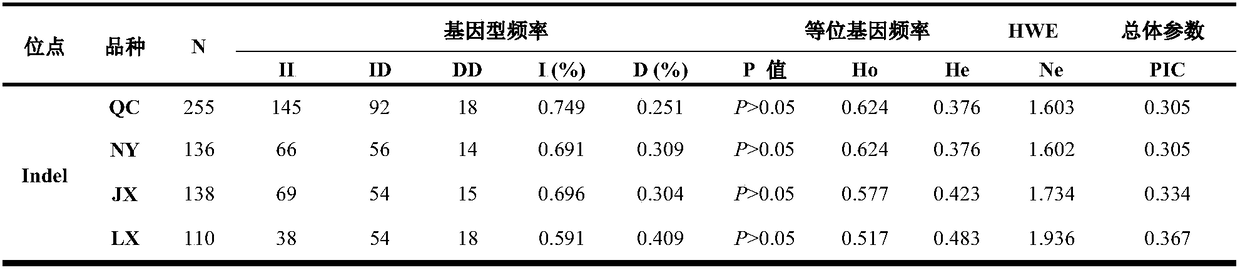
The invention discloses a detection method for 19-bp duplication and deletion polymorphism of a Chinese yellow cattle PLAG1 (Pleomorphic Adenoma Gene 1) gene and application of the detection method. The method takes Chinese yellow cattle whole genome DNA (Deoxyribonucleic Acid) as a template, and a primer pair P1 is used for treating a partial segment of the Chinese yellow cattle PLAG1 gene through PCR (Polymerase Chain Reaction) amplification; then the segment obtained by the PCR amplification is subjected to agarose gel electrophoresis; a gene type of an insertion / deletion polymorphism siteon the Chinese yellow cattle PLAG1 gene is identified according to an electrophoresis result; different gene types of the duplication and deletion polymorphism of the gene are remarkably related withgrowth properties including body height, body length, chest circumference, chest width, chest depth, waist height and the like four types of Chinese yellow cattle including Qiachuan cattle, Nanyang cattle, Jiaxian cattle and Luxi cattle. The method disclosed by the invention can be applied to marker-assisted selection breeding of growth characters of beef cattle, and a good genetic resource groupwith good growth performance is easily and rapidly established.
Owner:NORTHWEST A & F UNIV
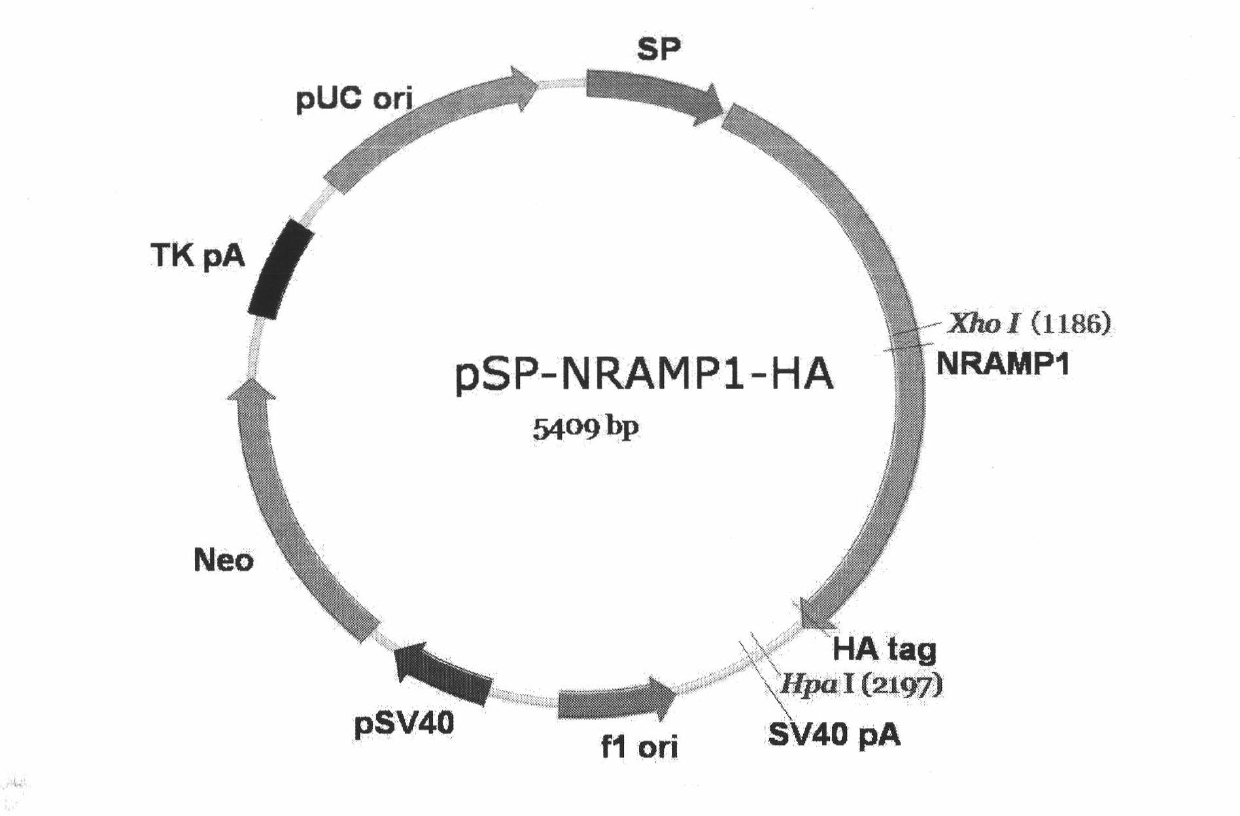
Integration cattle NRAMPl macrophage specificity expression vector/cattle fibroblast and method thereof
InactiveCN102559749AEfficient specificityPrevention of bovine brucellosisFermentationVector-based foreign material introductionCell systemBrucellosis
The invention discloses an integration NRAMPl macrophage specificity expression vector / cattle fibroblast and a method thereof. An expression vector pSP-NRAMPl-HA containing NRAMPl macrophage specificity and a Qinchuan cattle fibroblast system which is based on the vector and contains NRAMPl are structured, and the method utilizing the cell system as a nucleus donor cell provides a solid foundation for solving transgenosis cattle problem suffering from Qinchuan cattle brucellosis. According to the invention, the exogenous expression vector pSP-NRAMPl-HA is conducted into the cattle fibroblast by a liposome transfection method, negative cells are obtained by G418 screening, and proved by PCR, the aim gene NRAMPl is integrated into the genome of the cattle fibroblast; and finally the cattle fibroblast cell integrated with the NRAMP1 gene is taken as the nucleus donor, a nucleus-removing oocyte is moved into the nucleus donor, thus obtaining a transgenosis clone body.
Owner:NORTHWEST A & F UNIV
Plasmid type adenovirus vector pAd-NRIP1 and construction method thereof
InactiveCN102399819AEfficient expressionFermentationVector-based foreign material introductionDigestionLiposome
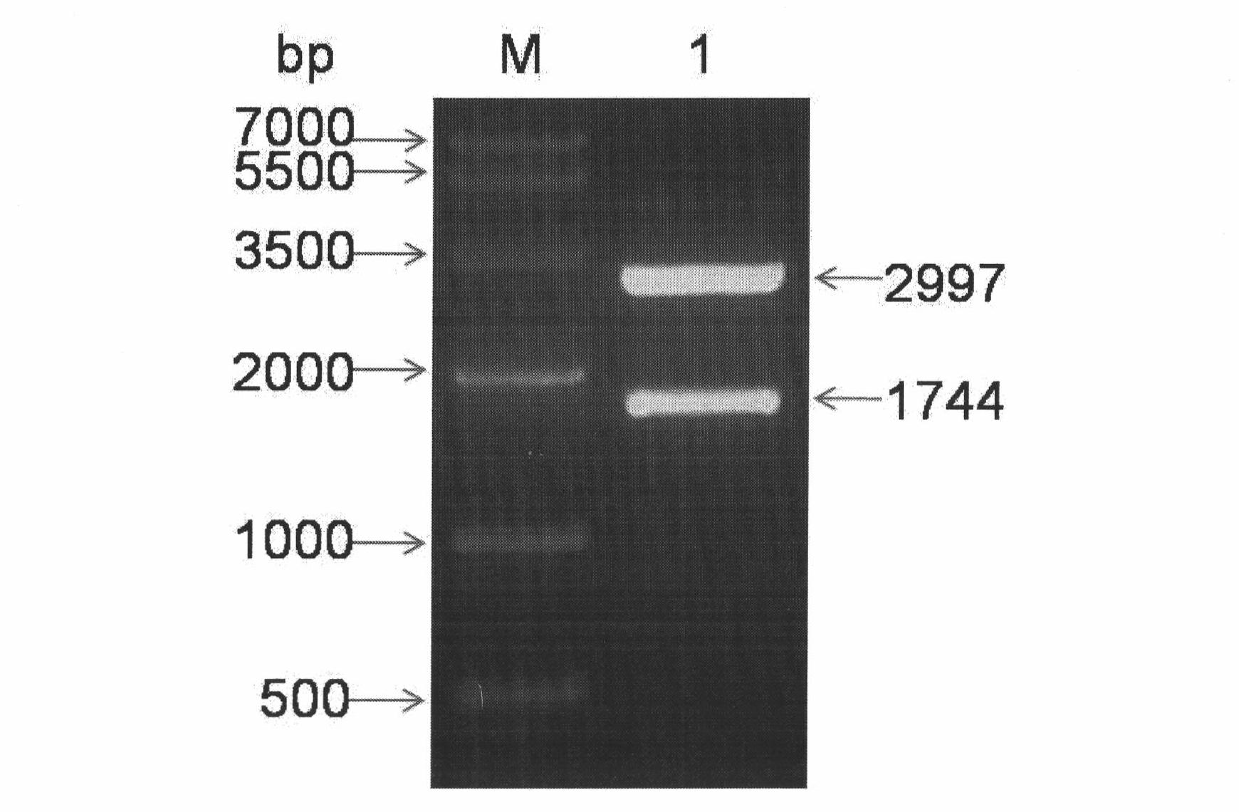
Construction of a recombinant adenovirus vector of an NRIP1 (nuclear receptor interacting protein 1) gene of Qinchuan cattle can be applied to gene function research and dentification on Qinchuan cattle NRIP1, reconstruction of seed cell, regulation on gene metabolism of muscle, fat and reproduction, etc. The Qinchuan cattle NRIP1 gene is inserted under a CMV promoter of a pAdTrack-CMV adenovirusshuttle plasmid to construct a recombinant shuttle plasmid pAdTrack-CMV-NRIP1. After linearization, a PmeI transforms an E.coli BJ5183 competent cell containing a pAdEasy-1 plasmid; an efficient homologous recombinant system of a Cre recombinase in the E.coli BJ5183 is utilized to complete homologous recombination of pAdTrack-CMV-NRIP1 and pAdEasy-1 in bacteria. A correct recombinant adenovirus plasmid is named as pAd-NRIP1. After digestion linearization of pAd-NRIP1 by a PacI, a liposome is transfected to a 293A cell, so as to obtain a recombinant virosome.
Owner:NORTHWEST A & F UNIV
Molecular marking method for indicating Qinchuan cattle meat tenderness and water-holding capacity by utilizing THRSP (thyroid hormone responsive spot) genes
InactiveCN103740830AFast improvementBroad development prospectsMicrobiological testing/measurementSingle-strand conformation polymorphismThyroid hormones
The invention relates to a molecular marking method for indicating Qinchuan cattle meat tenderness and water-holding capacity by utilizing THRSP (thyroid hormone responsive spot) genes. The method comprises the steps of designing a pair of primers according to a cattle THRSP gene sequence, executing the SSCP (single strand conformation polymorphism) band type detection on Qinchuan cattle whole-blood genomic group DNA (deoxyribonucleic acid) after PCR proliferation, and determining the Qinchuan cattle meat tenderness and the water-holding capacity by analyzing the band type. By adopting the PCR-SSCP technology and analyzing and comparing the variation point gene type, the meat quality of the cattle can be predicted and determined; the molecular marking method is simple to operate, low in expense, high in precision and capable of realizing rapid detection.
Owner:NORTHWEST A & F UNIV
Plasmid-type adenovirus vector pAd-PPARGC1A and construction method thereof
InactiveCN102766653AEnsure biosecurityEfficient expressionFermentationVector-based foreign material introductionLiposomeInverted Terminal Repeat
The invention relates to a plasmid-type adenovirus vector pAd-PPARGC1A (peroxisome proliferator-activated receptor gamma coactivator 1 alpha) and a construction method thereof, and belongs to the technical field of gene engineering. The construction method is as below: inserting a Qinchuan cattle PPARGC1A gene under a CMV promoter of a pAdTrack-CMV shuttle plasmid to construct a recombinant shuttle plasmid pAdTrack-CMV-PPARGC1A; linearizing by PmeI and transforming E.coli BJ5183 competent bacteria containing pAdEasy-l adenovirus backbone plasmids; carrying out homologous recombination on the pAdTrack-CMV-PPARGC1A and the pAdEasy-1 in bacteria, by using an efficient homologous recombination system of an E.coli BJ5183 endogenous Cre recombinase; naming a correct recombinant adenovirus plasmid after pAd-PPARGC1A; linearizing the pAd-PPARGC1A by Pac I and exposing inverted terminal repeats (ITR); and transfecting HEK-293A cells by the liposome to produce recombinant virus particles. The present invention has advantages of ensuring biological safety of the recombinant adenovirus vector, promoting efficient expression of the gene and benefiting expression of the gene for subsequent detection.
Owner:NORTHWEST A & F UNIV
Cattle Nramp1 macrophage specificity expression vector as well as construction method and application thereof
InactiveCN102226200ABoost expression levelUseful for detection of expression levelsVector-based foreign material introductionAnimal husbandryPhagocyteOpen reading frame
The invention relates to a cattle Nramp1 macrophage specificity expression vector as well as a construction method and application thereof. In the method, a plasmid pIRES2-EGFP (Internal Ribosome Entry Site-Enhanced Green Fluorescent Protein) serves as a skeleton carrier; a cattle Nramp1 open reading frame sequence is inserted between the multipe cloning site Sa1 I and BamH I; and the Nramp1 gene expression regulatory sequence is used for replacing a cytomegalovirus (CMV) on the skeleton carrier so as to realize the specificity expression of the Nramp1 gene in a phagocyte. The carrier can serve as a transgenic carrier to be applied in cultivating transgenic disease-resistant animals. The invention also points out that the similarity between the Qinchuan cattle Nramp1 open reading frame sequence and the corresponding sequence (of which the GenBank sequence number is U12862.1) in the GenBank is 99.88%, and two base differences exist.
Owner:NORTHWEST A & F UNIV
Method for rapidly detecting single nucleotide polymorphism (SNP) of bovine PNPLA3 (patatin like phospholipase domain-containing 3) gene and application of method
InactiveCN103805692AIncrease weightMicrobiological testing/measurementEnergy balancingEnzyme digestion
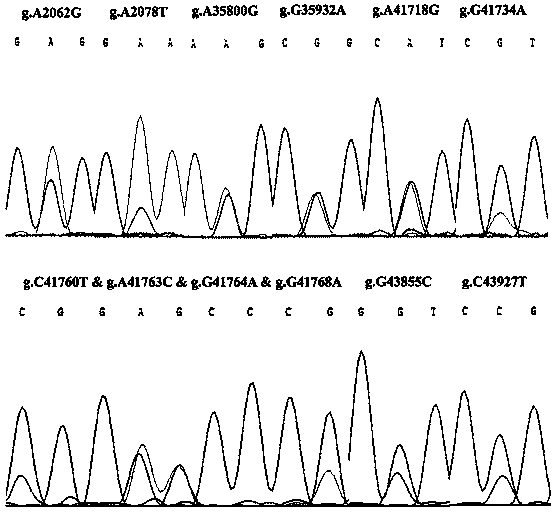
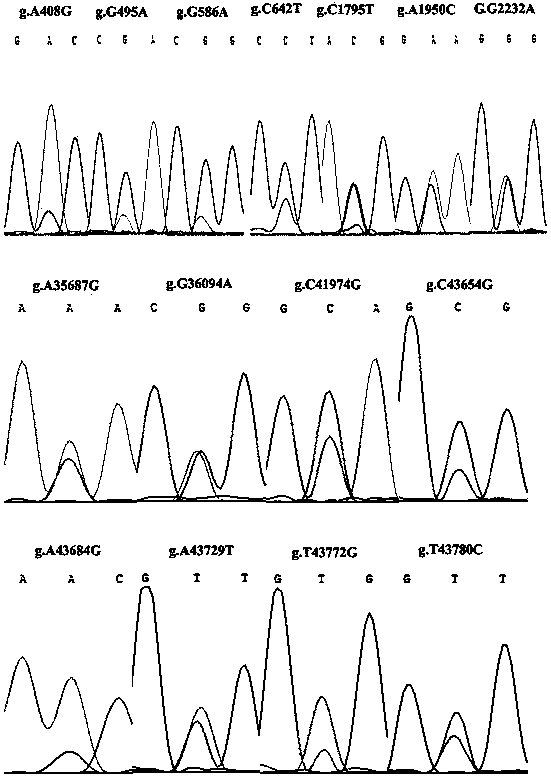
The invention discloses an RFLP (Restriction Fragment Length Polymorphism) method for rapidly detecting single nucleotide polymorphism (SNP) of bovine PNPLA3 (Patatin Like Phospholipase Domain-Containing 3) gene. The RFLP method comprises the steps of by taking PNPLA3 gene-containing to-be-detected whole-genome DNA as a template, and human-designed primer pairs P1-P5 as primers, carrying out PCR (Polymerase Chain Reaction) amplification on the bovine PNPLA3 gene, and discovering 27 SNP loci; selecting four missense mutation loci and respectively designing primer pairs P2-Bg1I, P2-RsaI, P3-BmgT120I and P3-MspI; carrying out PCR amplification by using the four primer pairs, respectively performing enzyme digestion to the PCR amplified product by using the four restriction endonuclease, and then detecting the enzyme digestion product by using agarose gel electrophoresis, wherein the electrophoresis result shows the SNP at the 2062th locus, the 2078th locus, the 35800th locus and the 35932th locus of the PNPLA3 gene of the Qinchuan bovine gene. The PNPLA3 gene relates to the growth traits of weight, average daily gain and the like, the PNPLA3 gene has the activity of lipase and acyltransferase and plays an important role in aspects of maintaining energy balance, the detection method can be used for marker assisted selection (MAS) breeding of the growth traits of China cattle, thus being beneficial to fast setting cattle population with excellent genetic resources.
Owner:NORTHWEST A & F UNIV
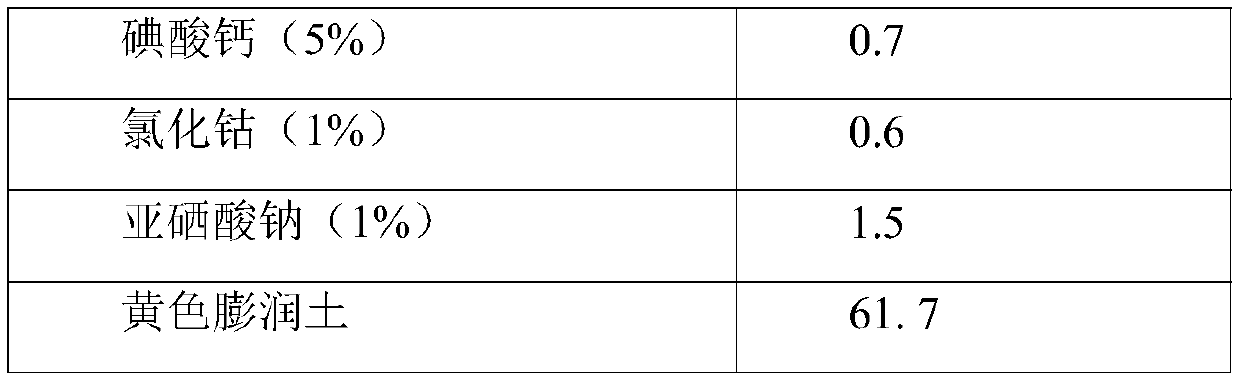
Fodder for improving beef quality of Qinchuan cattle and preparation method of fodder
InactiveCN109984262AImprove conversion rateNutritional balanceFood processingAnimal feeding stuffRumenMonensin
The invention discloses a fodder for improving beef quality of Qinchuan cattle and a preparation method of the fodder. The fodder contains corn, soya bean meal, cottonseed meal, corn alcohol grains, wheat bran, apple residues, baking soda, salt, calcium hydrophosphate, mountain flour, Chinese herbal medicine mixtures, rumen-protected leucine, a microelement additive, multivitamins, a saccharomycescerevisiae culture and monensin. According to the fodder, all the raw materials are subjected to reasonable compatibility, the rumen-protected leucine and the Chinese herbal medicine mixtures are added, the health of the rumen intestinal tracts of the Qinchuan cattle is adjusted synergistically, and the immunity of the cattle is enhanced; sufficient balanced nutrition is provided, rapid weight increment of the cattle is promoted, the color, tenderness and water-holding capacity of Qinchuan beef are improved, and the beef quality is improved.
Owner:西安铁骑力士饲料有限公司
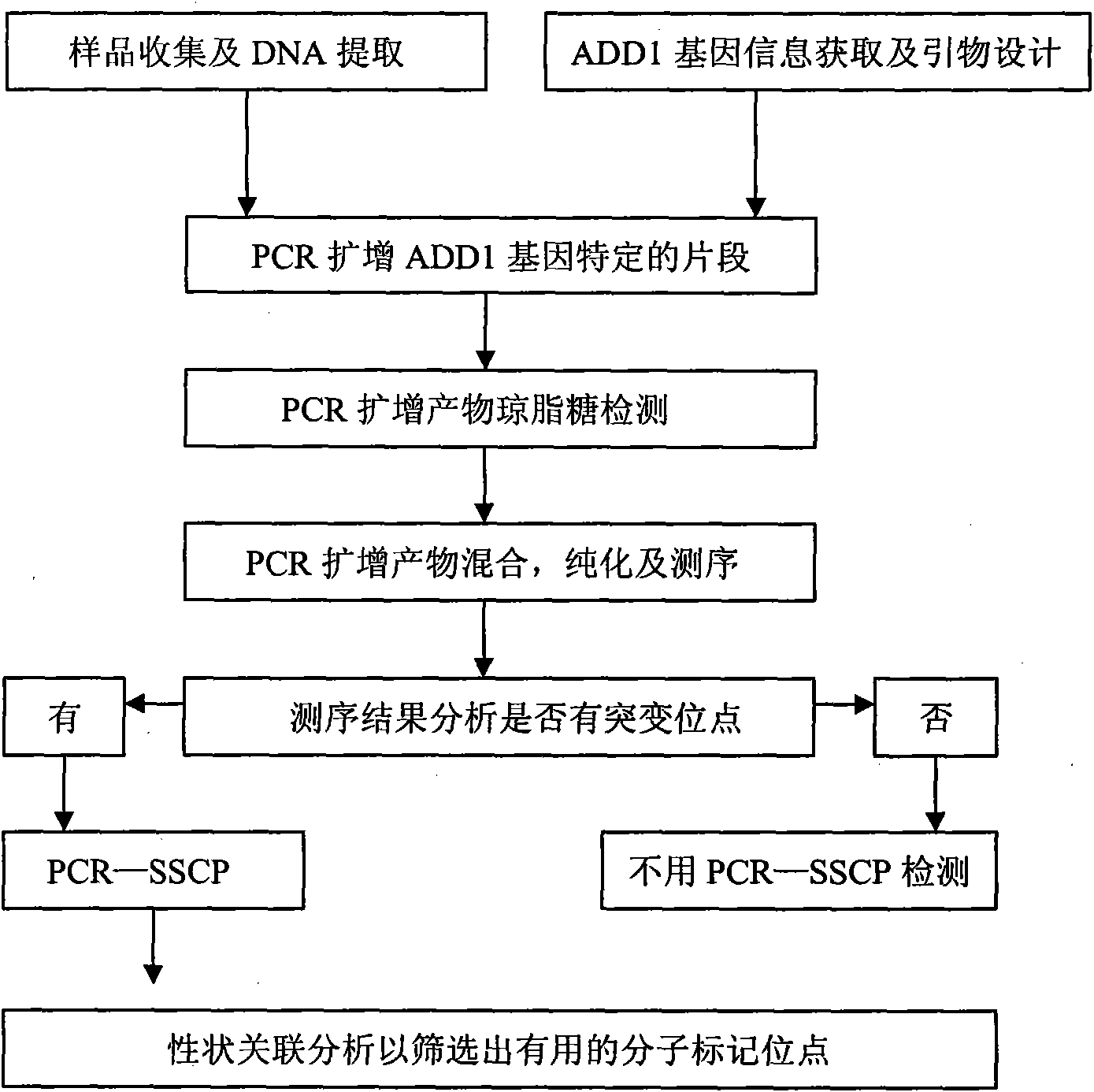
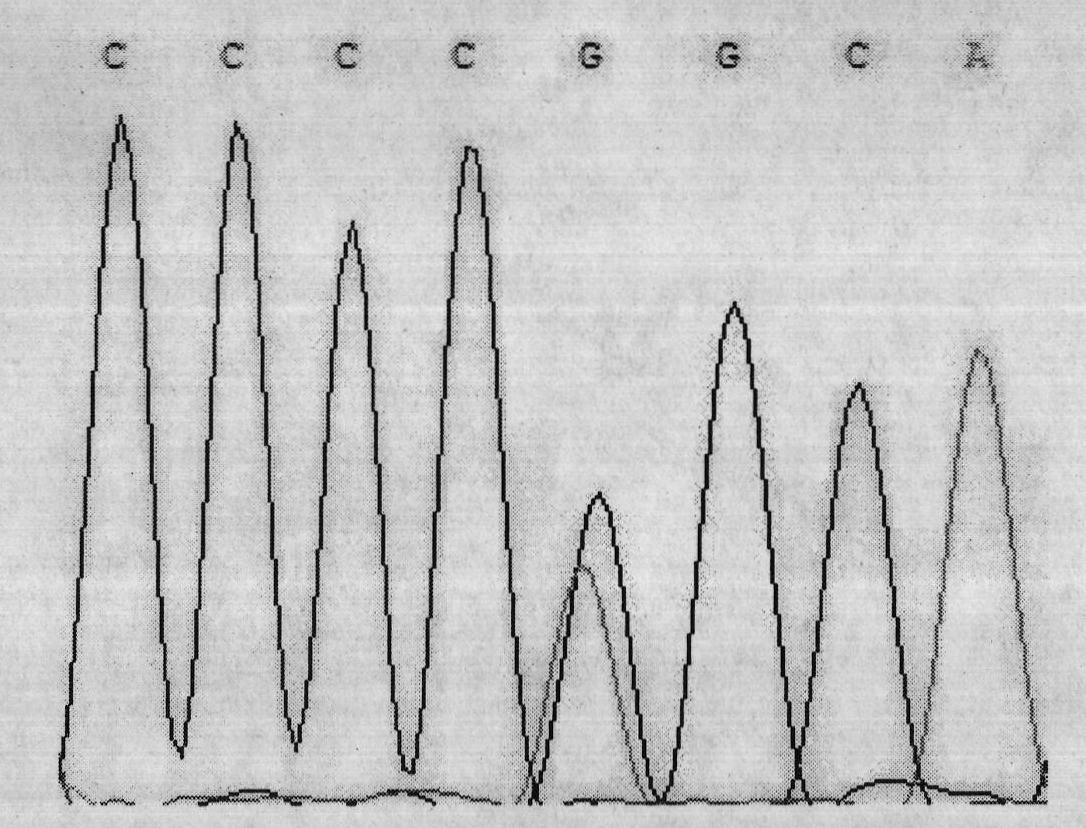
Single nucleotide polymorphism (SNP) of ADD1 gene of cattle and detection method thereof
InactiveCN102031304AAvoid instabilityLeak detectionMicrobiological testing/measurementSingle-strand conformation polymorphismGenomic DNA
The invention discloses a method for detecting the single nucleotide polymorphism (SNP) of an ADD1 gene of cattle, which comprises the following steps: through taking a genomic DNA of cattle containing an ADD1 gene as a template and taking a primer mix P as a primer, the ADD1 gene of cattle is amplified through PCR (polymerase chain reaction), then the segment size of the obtained product is amplified through AGE (agarose gel electrophoresis) detection; and then, the detection on SSCP (single strand conformation polymorphism) is performed. The polymorphism of the gene comprises a base polymorphism that the 70027th bit in an ADD1 gene sequence table SEQID No.8 of cattle is G or A, and a base polymorphism that the 70051th bit in the ADD1 gene sequence table SEQID No.8 of cattle is G or A. The analysis on the correlation between the SSCP polymorphism and the production performance shows that: a certain correlation exits between the polymorphic site and the production performance, therefore, the weight of a Qinchuan cattle (genotype: BB) is significantly higher than that of a cattle (genotype: AA). The method is used for the auxiliary selection and molecular breeding of cattle through screening and detecting molecular genetic markers closely relating to the growth properties of cattle on a DNA level, therefore, the method plays a vital role in improving the production performance of cattle.
Owner:XUZHOU NORMAL UNIVERSITY
BBS2 molecular marker associated with beef quality traits of beef cattle, and detection kit thereof
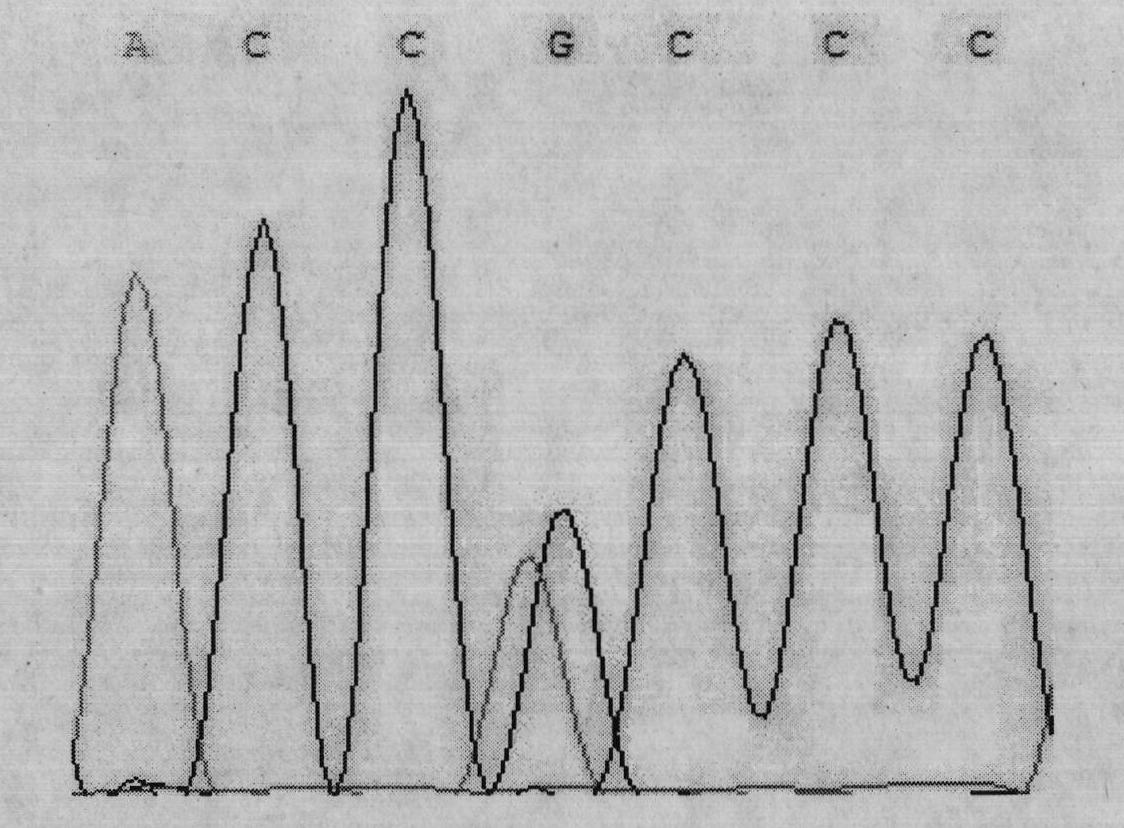
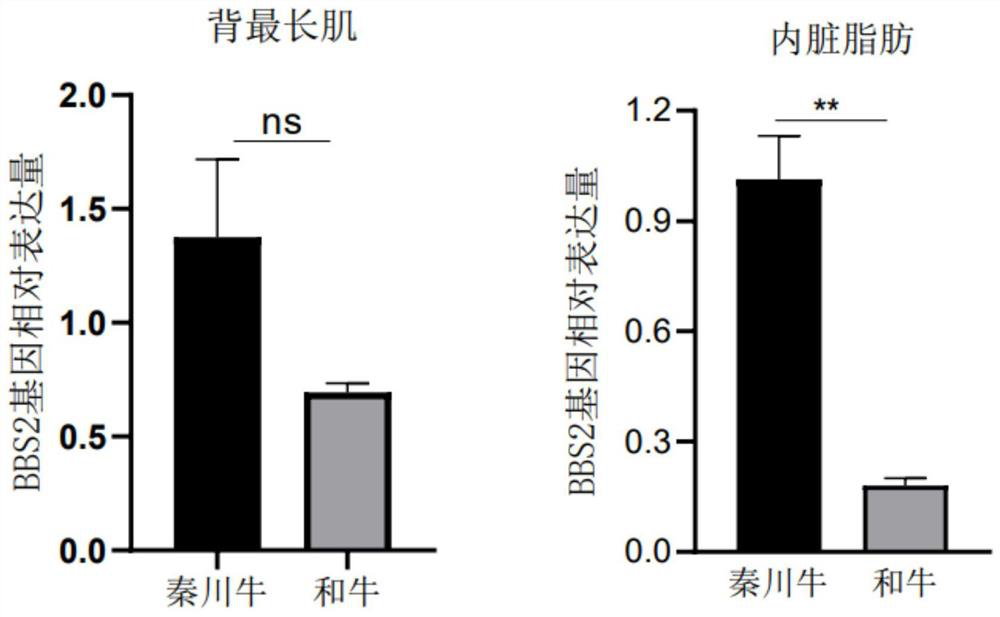
The invention discloses a BBS2 molecular marker associated with beef quality traits of beef cattle, a detection kit and application of the BBS2 molecular marker. The molecular marker is located at a g.24223562G / A site, a g.24226239C / T site and a g.24227851G / A site of a BBS2 gene coding region, and the molecular marker is remarkably related to beef cattle eye muscle area, eye muscle depth and intermuscular fat content meat quality traits. The marker is a potential breeding genetic marker for local yellow cattle meat represented by Qinchuan cattle. Based on the discovery, the kit for rapidly detecting the BBS2 genotype to predict the beef quality traits of the beef cattle is developed, and the kit can be used for early breeding of local cattle represented by Qinchuan cattle, the genetic progress of the beef cattle is accelerated, and the production cost is saved. The beef cattle early-stage selection and reservation are realized, and the beef breeding process is accelerated.
Owner:NORTHWEST A & F UNIV
Method for detecting linked mutation of AMPD1 gene of Qinchuan cattle
The invention discloses a method for detecting the linked mutation of an AMPD1 gene of Qinchuan cattle. The method is characterized in that: a complete genomic DNA sequence of Qinchuan cattle to be measured and containing the AMPD1 gene is used as a template; polymerase chain reaction primers are designed under the presence of 2*Reaction Mix, ddH2O, a Forward primer, a Reverse primer, a Golden DNA polymerase, and a DNA template and are amplified under a PCR reaction program; PCR products are degenerated after the amplification; and amplified fragments are subjected to polyacrylamide gel electrophoresis and are analyzed through a gel imaging system to determine single nucleotide polymorphisms (SNPs) of the AMPD1 gene. The detection method has the advantages of simple operation, rapidness, low cost, high detection accuracy and convenient popularization and application.
Owner:NORTHWEST A & F UNIV
Detection method of single nucleotide polymorphism of Qinchuan cattle microRNA-320a-1 gene and application thereof
ActiveCN106521001AAccurate detectionQuick checkMicrobiological testing/measurementEnzyme digestionMarker-assisted selection
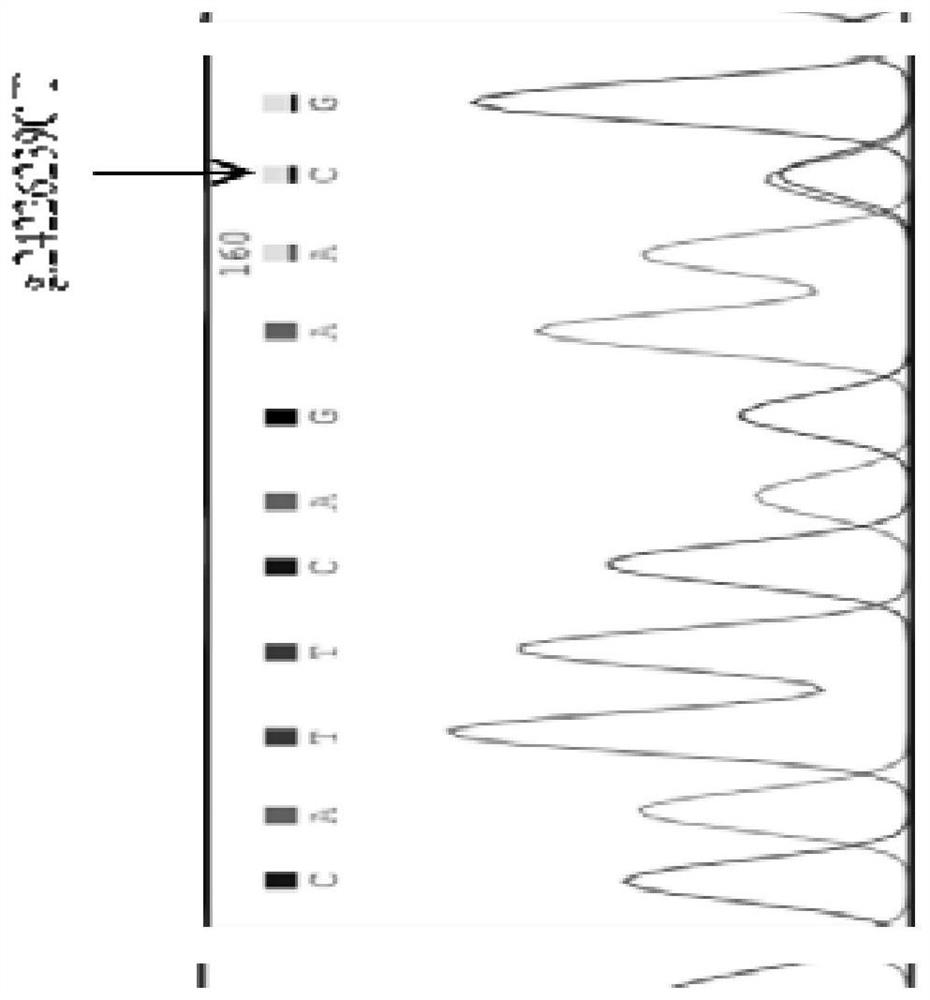
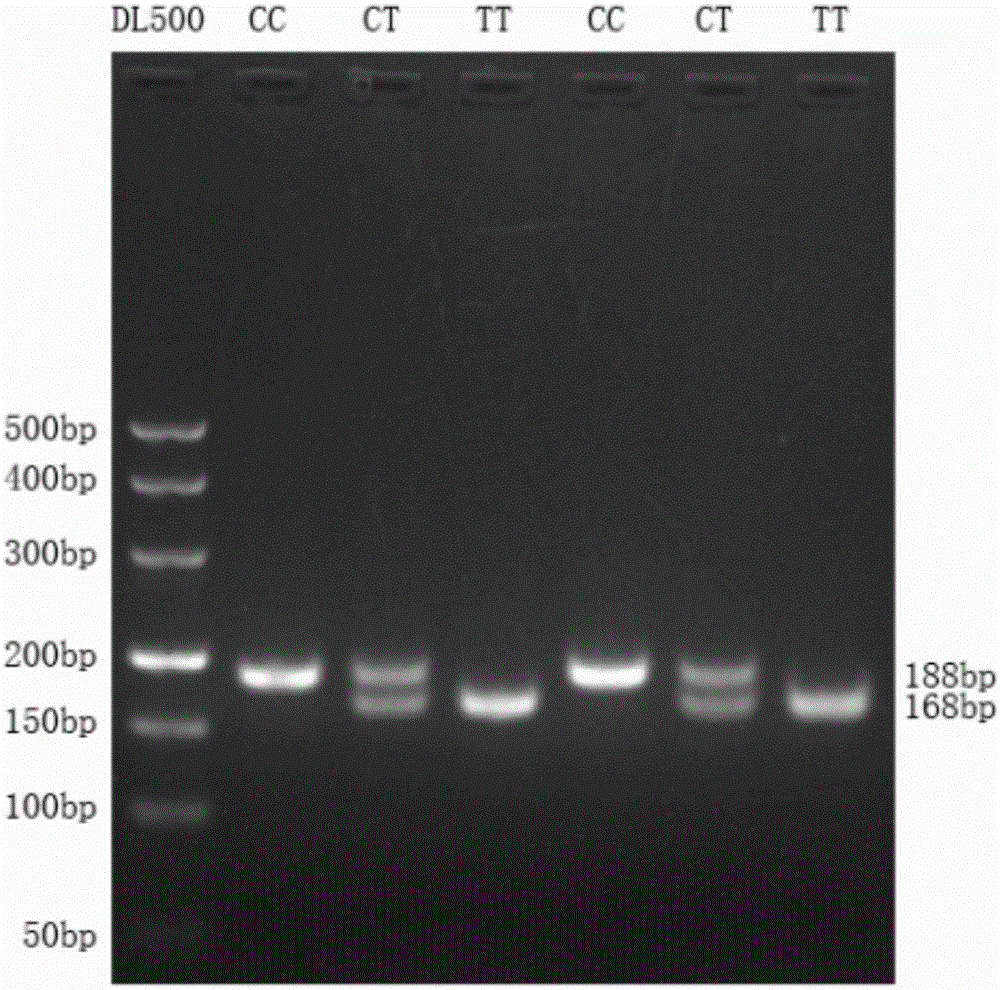
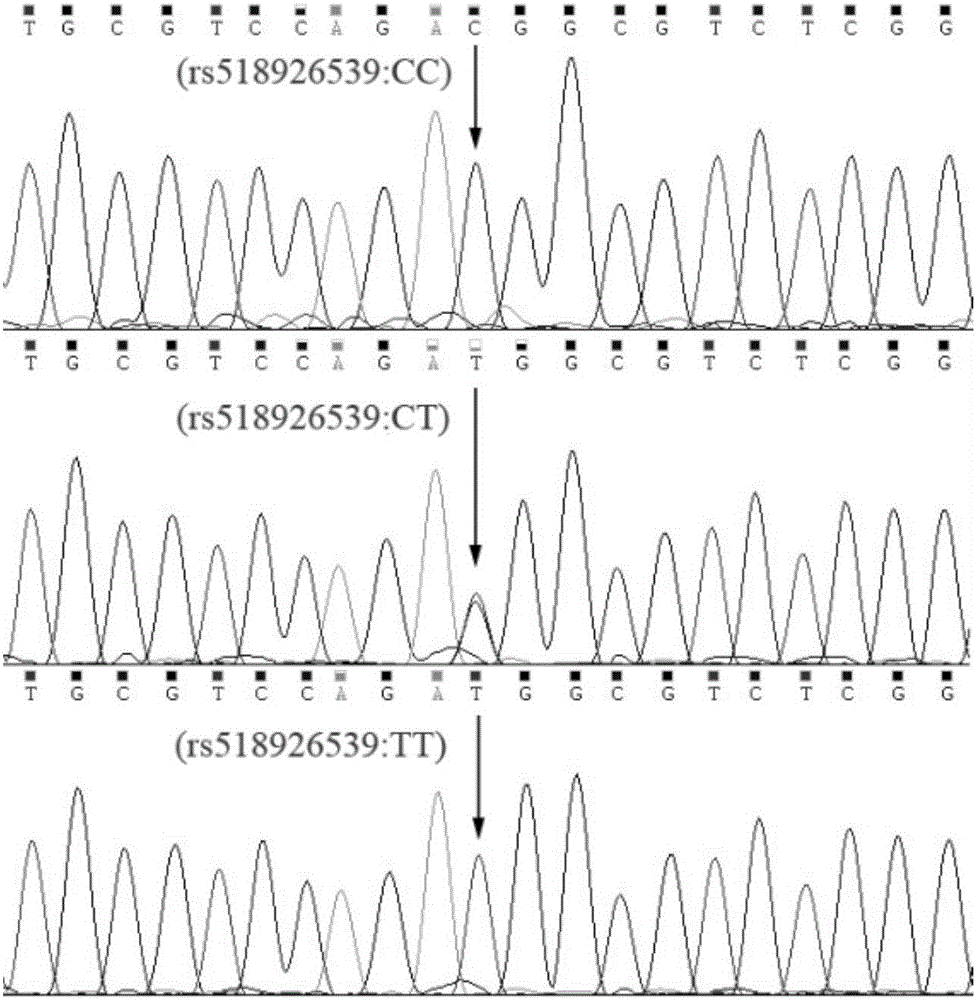
The invention discloses a detection method of single nucleotide polymorphism of a Qinchuan cattle microRNA-320a-1 gene and an application thereof. The detection method comprises the following steps: with the to-be-detected Qinchuan cattle whole-genome DNA as a template and a primer pair P2 as primers, performing PCR amplification of a Qinchuan cattle microRNA-320a-1 gene segment; digesting the PCR amplification product with restriction endonuclease PuvII, and performing agarose gel electrophoresis of the amplified segment after the enzyme digestion; and identifying the single nucleotide polymorphism of Qinchuan cattle microRNA-320a-1 gene rs518926539:C>T according to the result of agarose gel electrophoresis. The detection method disclosed by the invention lays a foundation for the marker assisted selection of the growth character for Qinchuan cattle meat based on the relationship between the microRNA-320a-1 gene SNP and the growth character, and is favorable for quickly establishing a Qinchuan cattle population with good genetic resources.
Owner:NORTHWEST A & F UNIV
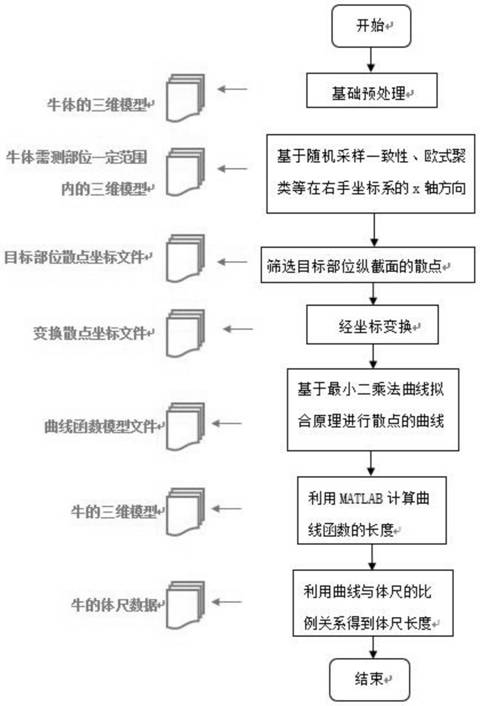
Method for measuring body size of living Qinchuan cattle based on 3D camera
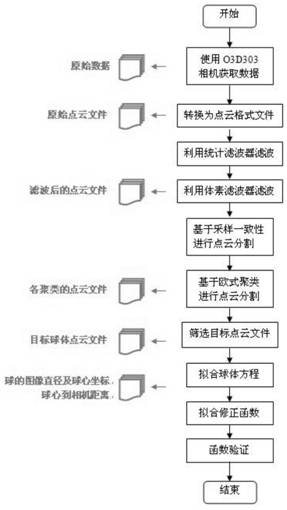
PendingCN114429497ATo achieve the purpose of body measurementAchieve the purpose of measurementImage enhancementImage analysisRadiologyBody height
The invention discloses a method for measuring the body size of a living Qinchuan cattle based on a 3D camera. Indoor regular object point cloud images are obtained through a 3D laser camera, and by means of filtering, segmentation, curve fitting, correction and other algorithms, through coordinate transformation and by means of a space curve length measurement method, the non-contact measurement function of 11 body sizes, namely, the body height, the waist height, the body slope length, the jijiang length, the chest width, the jijiang width, the chest circumference, the abdominal circumference, the tube circumference, the hip width and the waist angle width, of a target cattle body is finally achieved. The problems of high difficulty and low efficiency of manual measurement of the cattle body size are solved to a certain extent.
Owner:NORTHWEST A & F UNIV
Method for detecting body sizes of Qinchuan cattle through using ANAPC13 gene
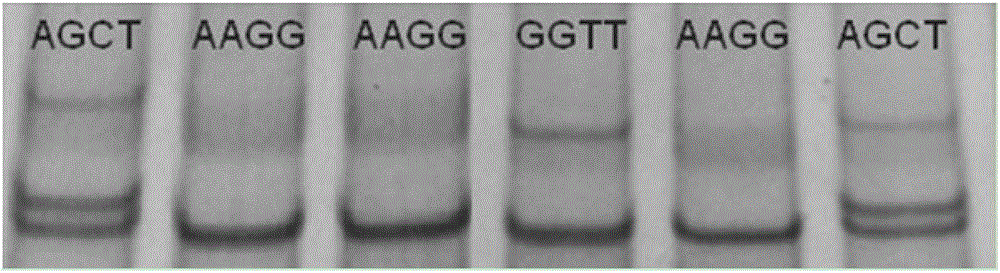
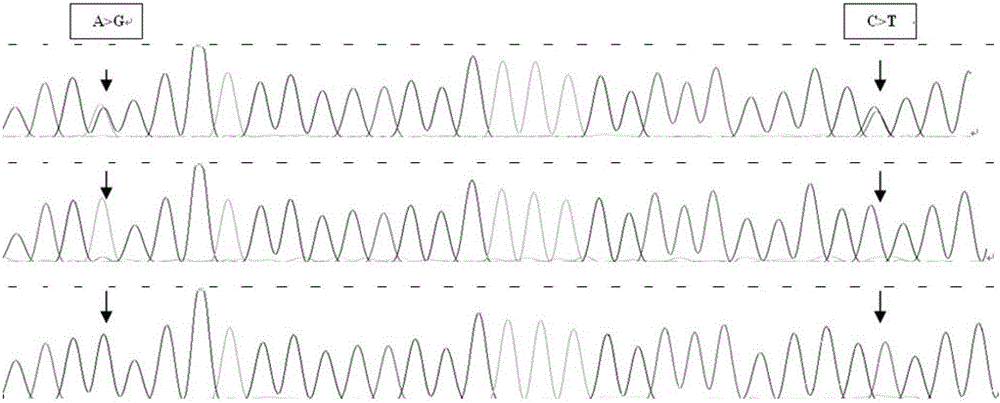
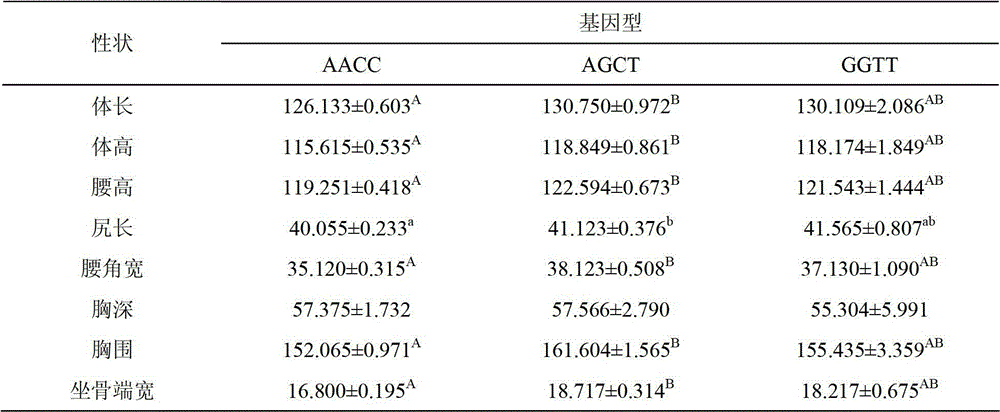
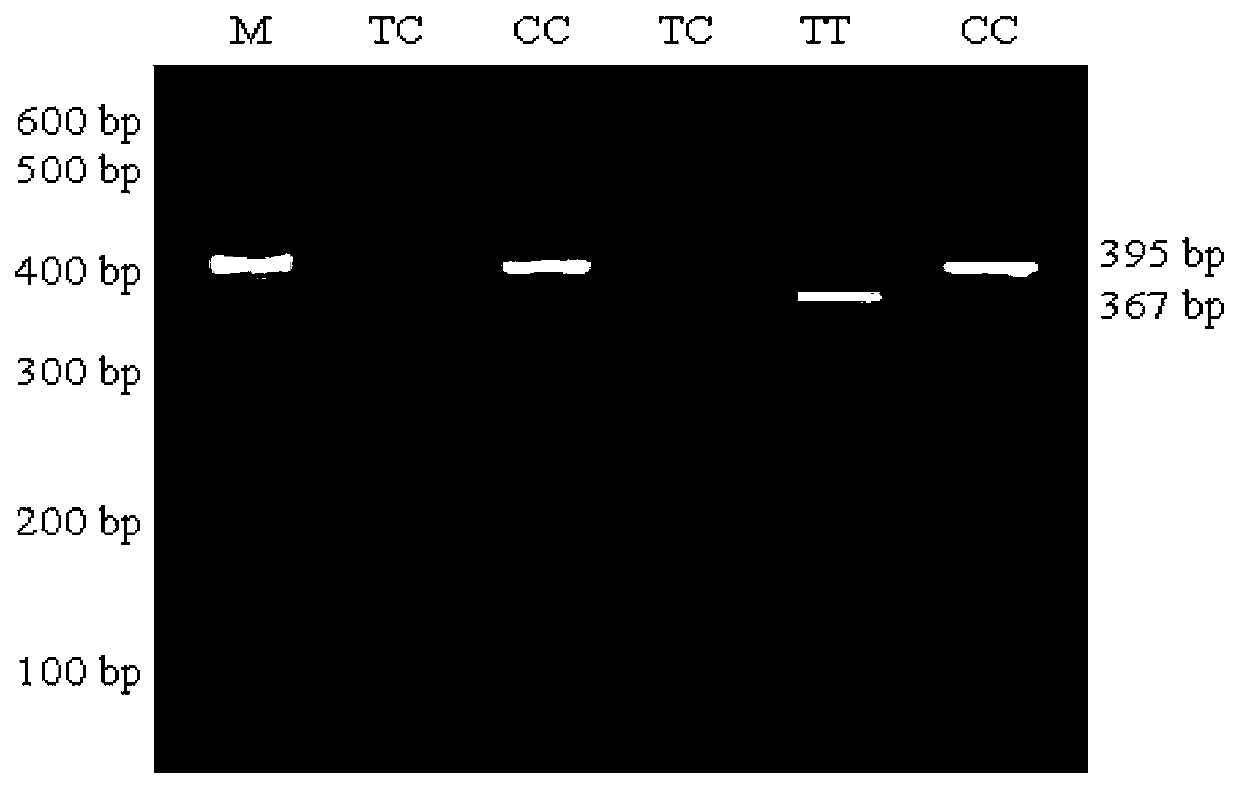
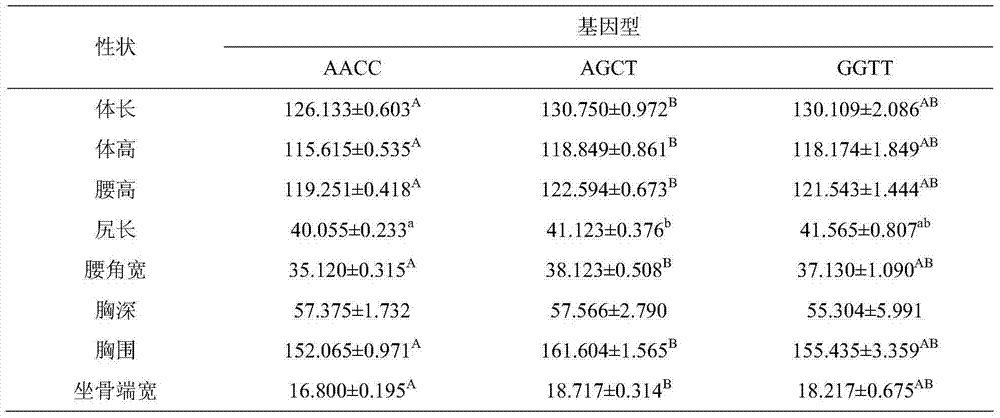
ActiveCN102747150AEasy to operateLow costMicrobiological testing/measurementMolecular breedingGenotype
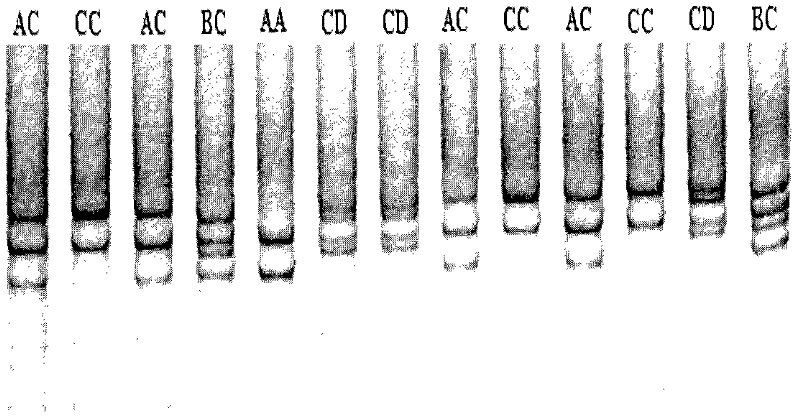
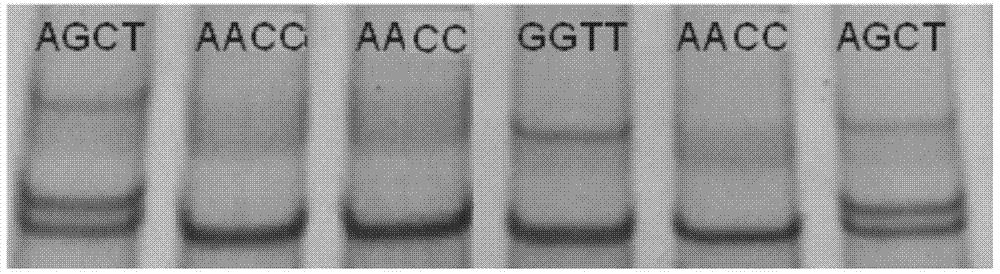
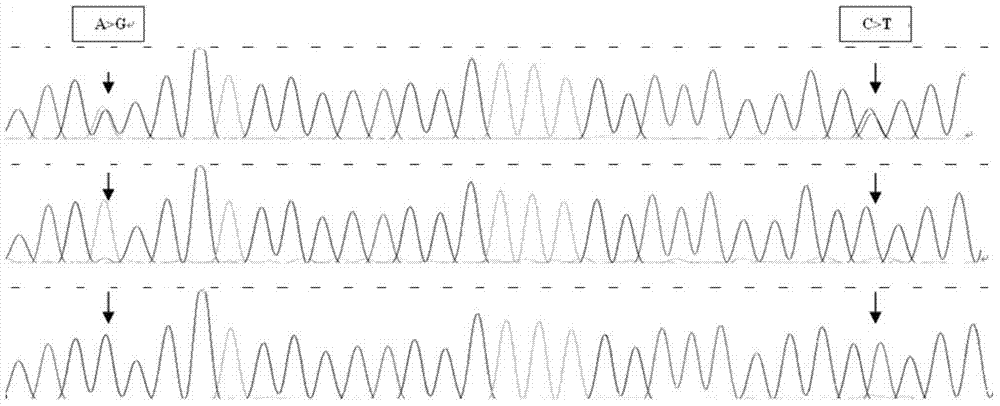
The invention discloses a method for detecting the body sizes of Qinchuan cattle through using ANAPC13 gene. The method adopts the combination of a PCR-SSCR technology with DNA sequencing to detect the variation sites of the ANAPC13 gene of the cattle. An SSCP result shows that there are three banding patterns, and a sequencing result shows that there are two variation sites comprising an A-G mutation at 17bp of a first exon and a C-T mutation at 42bp of the first exon. Later sequencing verification shows that the mutations of the two sites in 98% of individuals belong to a linkage mutation. So the three banding patterns are named as AACC, AGCT and GGTT respectively, and the Qinchuan cattle with the number being 404 undergo parting statistics and association analysis of the parting statistics with the body size character. The method is used for auxiliary selection and molecular breeding of the Qinchuan cattle to accelerate the breeding speed of fine breeds of the Qinchuan cattle through screening and detecting molecular genetic markers closely related with the growth and development traits of the Qinchuan cattle in DNA level. The body sizes of the Qinchuan cattle are predicted and determined through the analytic comparison of the genotype, so the method has the advantages of simple operation, low cost, high accuracy, and rapid detection.
Owner:NORTHWEST A & F UNIV
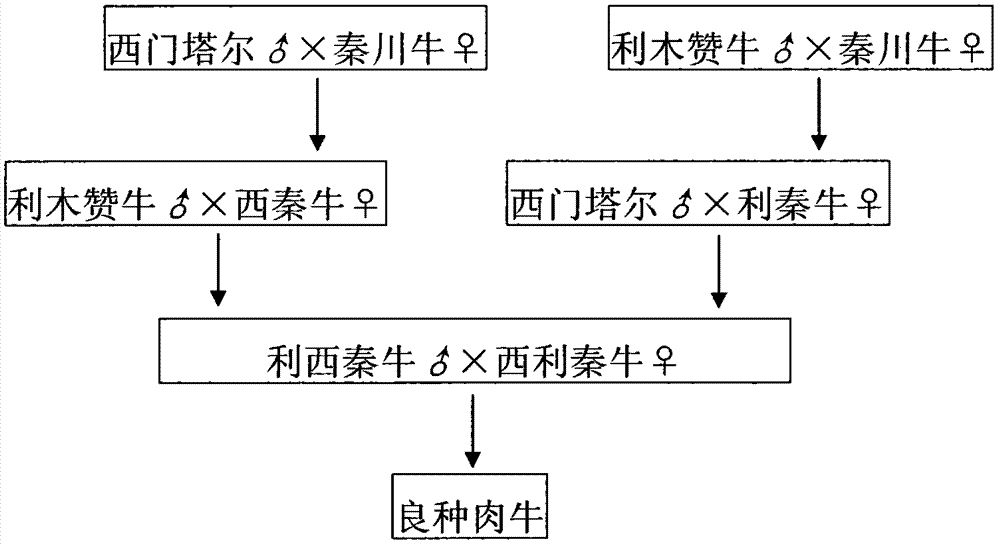
Breeding method for fine-breed beef cattle
InactiveCN104255654ASolve quality problemsSmall amount of solutionAnimal husbandrySimmental cowAgricultural science
The invention provides a breeding method for fine-breed beef cattle, and relates to the technical field of breeding methods. The breeding method comprises the following steps of: 1) hybridizing a male parent of Simmental with a female parent of Qinchuan cattle to generation F1, that is, Simmental-Qinchuan cattle; 2) hybridizing a male parent of limousine cattle with a female parent of Qinchuan cattle to generation F1, that is, limousine-Qinchuan cattle; 3) hybridizing a male parent of limousine cattle with a female parent of Simmental-Qinchuan cattle to generation F2, that is, limousine-Simmental-Qinchuan cattle; 4) hybridizing a male parent of Simmental with a female parent of limousine-Qinchuan cattle to generation F1, that is, Simmental-limousine-Qinchuan cattle; 5) hybridizing a male parent of limousine-Simmental-Qinchuan cattle with a female parent of Simmental-limousine-Qinchuan cattle to obtain the fine-breed beef cattle. According to the breeding method provided by the invention, the excellent characteristics of limousine cattle and Simmental cattle are inherited through crossbreeding on the basis of the excellent characters of Qinchuan cattle, thus achieving the production performance level of modern high-grade beef cattle varieties, solving the problems of low production quality, low amount and low efficiency of beef cattle products in China, and realizing economic character combination.
Owner:重庆东奇畜禽养殖有限公司
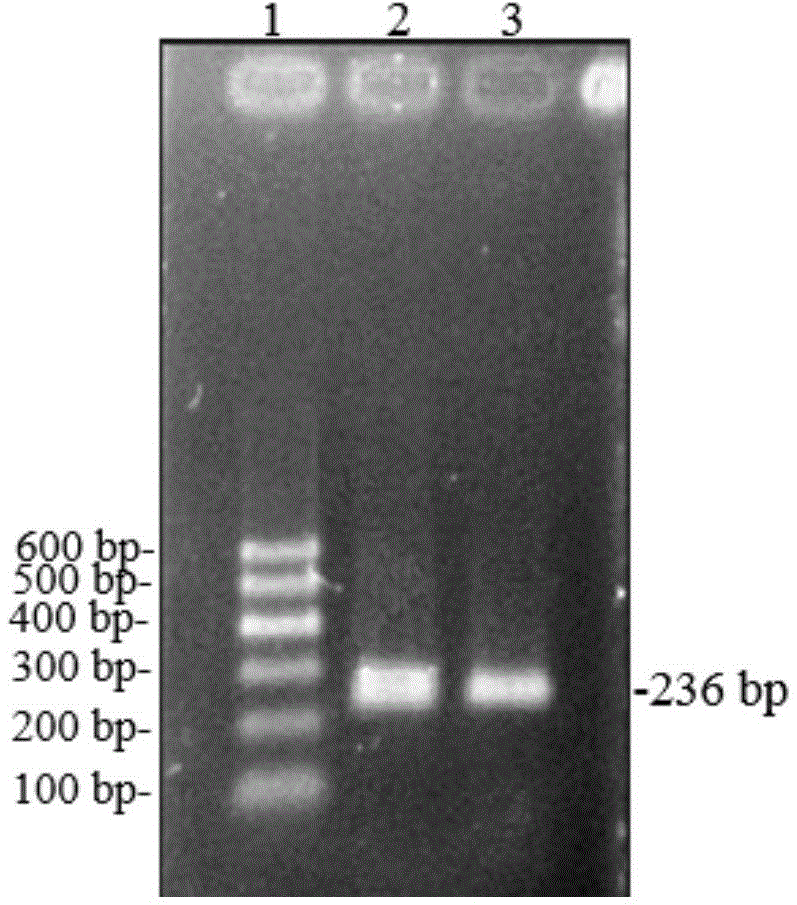
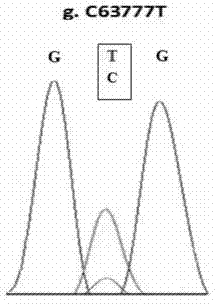
SNP molecular marker for CFL1 gene of Qinchuan cattle and detection method of SNP molecular marker
ActiveCN110564867AExcellent genetic resourcesAccurately estimate breeding valuesMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionElectrophoresis
The invention belongs to the field of molecular genetics, and provides an SNP molecular marker for CF1 gene of Qinchuan cattle and a detection method of the SNP molecular marker. The whole genome DNAof Qinchuan cattle to be tested containing the CFL1 gene is used as a template, a primer pair P is used as primers, and PCR is carried out to amplify the CFL1 gene; agarose gel electrophoresis is performed on the amplified fragments after enzyme digestion, after the PCR amplified product is digested with a restriction enzyme HinfI; and the base polymorphism at position 2052 of the CFL1 gene of Qinchuan cattle is identified according to the electrophoresis results. Because the mutation site is closely related to the beef traits (body length, chest width, chest depth and weight) of Qinchuan cattle, and the method is a method for screening and detecting the molecular genetic markers closely related to the growth traits of Qinchuan cattle at DNA level, the SNP molecular marker and the method can be used in auxiliary selection and molecular breeding of Qinchuan cattle, so as to accelerate the breeding speed of Qinchuan cattle.
Owner:YANGZHOU UNIV
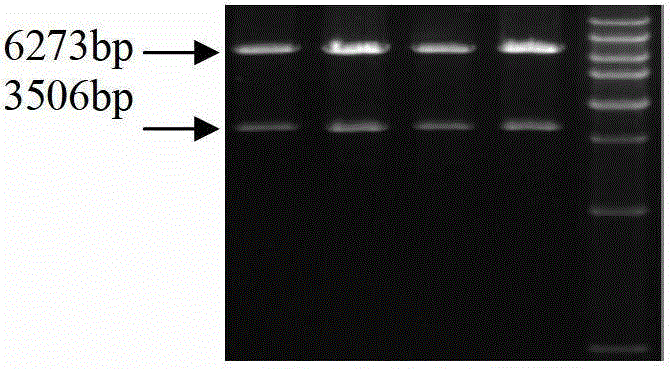
Method for detecting GAL3ST1 gene copy number variation of cattle and application thereof
ActiveCN111394474AAccurate identificationLow costMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyGenome
The invention discloses a method for detecting copy number variation of a GAL3ST1 gene of cattle and application thereof. According to the method, based on real-time quantitative PCR, the DNA of a genome of cattle (such as Qinchuan cattle) is used as a template, partial fragments of the copy number variation region of the GAL3ST1 gene of cattle are amplified by utilizing a pair of specific PCR primers, partial fragments of a universal transcription factor 3 gene of the cattle are used as internal reference by utilizing another pair of specific PCR primers, the quantitative result is calculatedby utilizing a method of 2*2-delta delta Ct, and the copy number variation type of an individual is determined. The method is established on the correlation basis between the copy number variation and growth traits of the GAL3ST1 gene of cattle and is favorable to accelerating the cattle molecular mark assisted selective breeding work. The method is simple and quick and is convenient for popularization and application.
Owner:NORTHWEST A & F UNIV
Preparation method of dry powder fermented mixed cattle feed
InactiveCN107873971AFully absorbedFully decomposedFood processingAnimal feeding stuffBiotechnologyFiber
The invention discloses a feed for feeding Qinchuan cattle. The feed is prepared by the following steps: (1) preparing a fermenting raw material feed bag; (2) preparing a protein raw material feed bag; (3) preparing a fiber green feed; (4) putting the fermenting raw material feed bag obtained in the step (S1), the protein raw material feed bag obtained in the step (S2) and the fiber green feed obtained in the step (S3) into a stirrer, and adequately stirring, so as to obtain a feeding feed mixture; (S5) dehydrating the feed mixture obtained in the step (S4), wherein the feed mixture is a mixture formed by mixing a fermenting raw material component, a protein raw material component and a green feed component in a mass ratio of 3 to 2 to 5. By mixing fermented difficultly-digested raw material components with proteins, microelements and nutrient substances, the adequate absorption and decomposition of the cattle to the raw materials can be greatly promoted, and the difficultly-digested raw material components can be converted into nutrients which can be directly absorbed by the cattle by adequately utilizing the autolysis of microorganisms and probiotics, so that the utilization rateof the feed is greatly increased.
Owner:CHENGDU XIANXIANXIAN BIOTECH CO LTD
Cattle serine/threonine kinase STK11 interfering adenovirus construction method
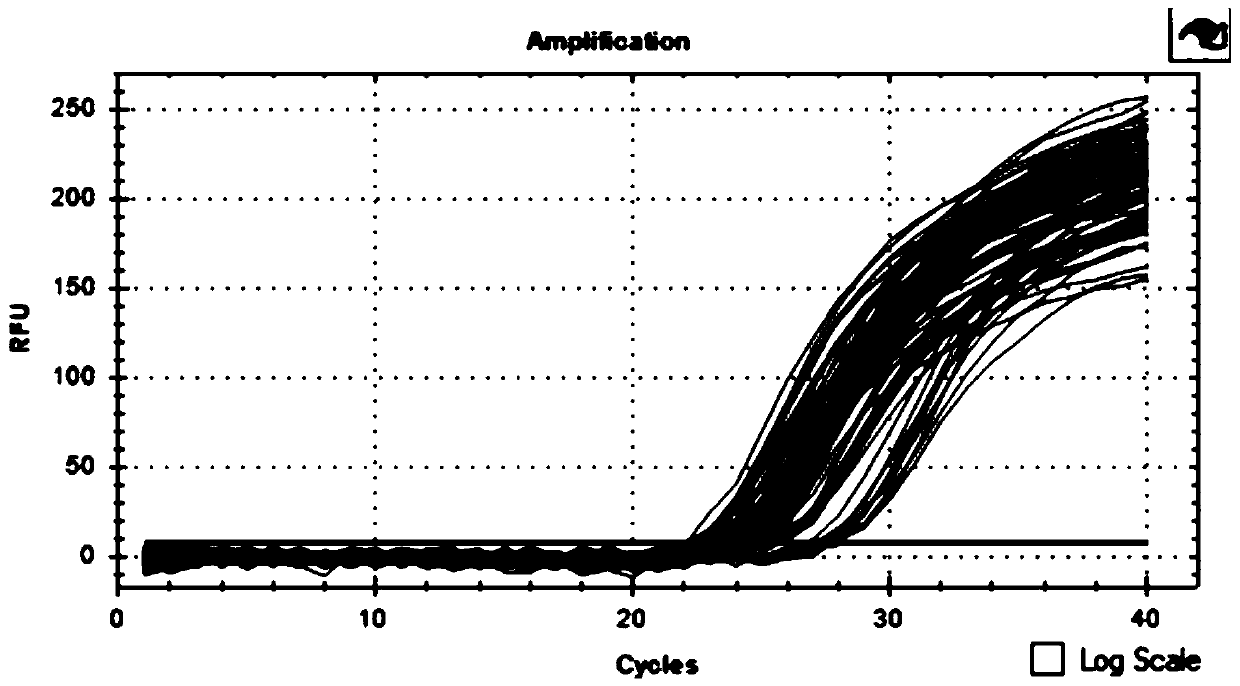
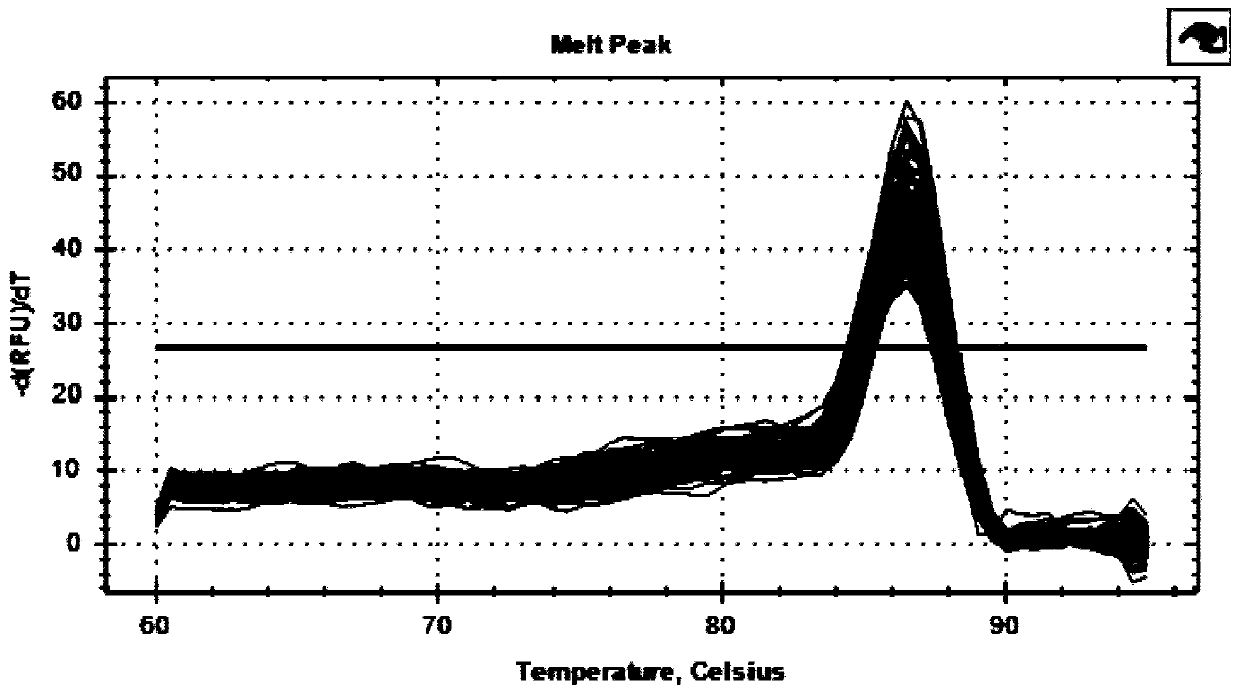
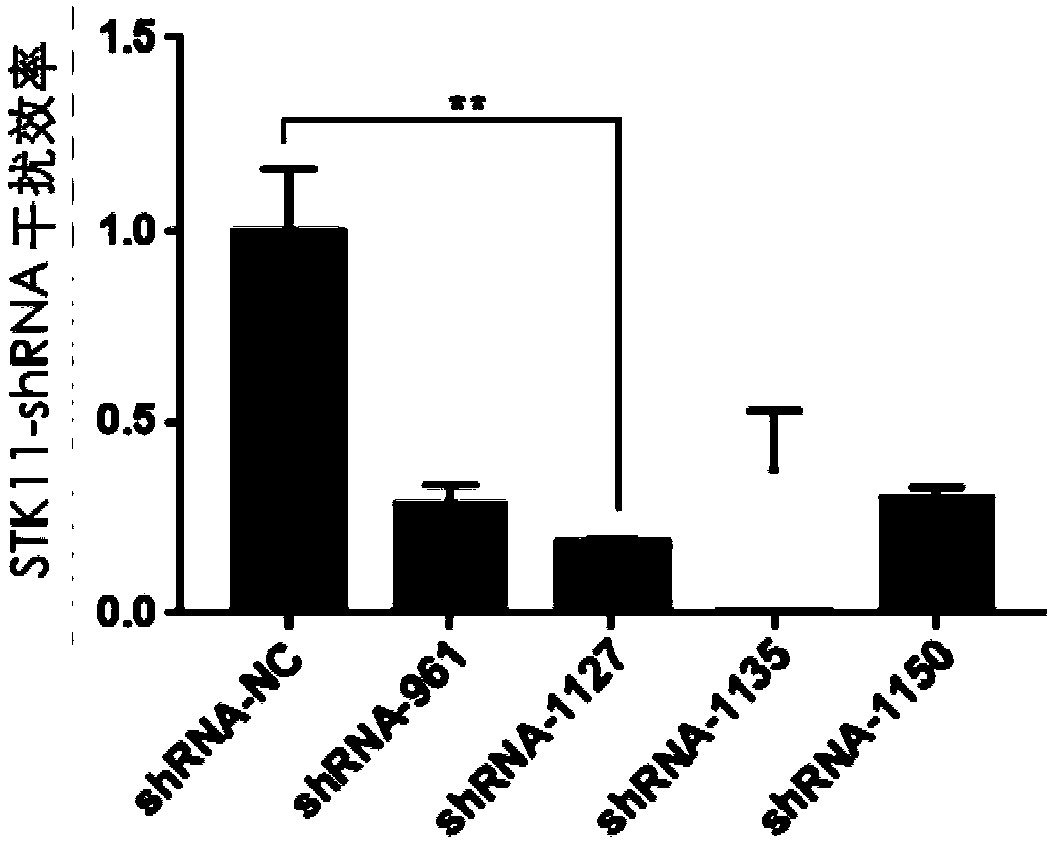
InactiveCN109593785AAchieving sustained knockdownInfection effect is goodNucleic acid vectorFermentationThreonineKinase
The invention relates to a cattle serine / threonine kinase STK11 interfering adenovirus construction method, an shRNA sequence with the optimal interfering efficiency is firstly screened, then the shRNA sequence is recombined with an interfering adenovirus vector to construct the STK11 interfering adenovirus; the infecting and the interfering efficiency of the STK11 interfering adenovirus are verified in Qinchuan cattle primary preadipocyte, the result shows that the infecting effect is good, the expression of the STK11 on the mRNA level is reduced by 76%, the protein level is also remarkably reduced, continuously knocking down an STK11 gene in a somatic cell is successfully realized, and the reliable experiment technique is provided for conducting the related researches on the function ofthe STK11 gene in future.
Owner:NORTHWEST A & F UNIV
Cattle plasmid adenovirus vector pAd-ZBED6 as well as construction method and use of cattle plasmid adenovirus vector pAd-ZBED6
InactiveCN103031336AMicrobiological testing/measurementBiological testingAgricultural scienceTyrosine recombinase
The invention relates to a cattle plasmid adenovirus vector pAd-ZBED6 as well as a construction method and use of the cattle plasmid adenovirus vector pAd-ZBED6, which belongs to the technical field of gene engineering. The construction method of the cattle plasmid adenovirus vector pAd-ZBED6 comprises the follow steps: (1) inserting Qinchuan cattle ZBED6 gene into a CMV (cytomegalovirus) promoter of a pAdTrack-CMV adenovirus shuttle plasmid to construct a recombinant shuttle plasmid pAdTrack-CMV-ZBED6; (2) after linearization of Pme I, converting an E.coil BJ5183 competent cell containing a pAdEasy-1 plasmid, completing homologous recombination of pAdTrack-CMV-ZBED6 and pAdEasy-1 in bacteria by means of an efficient homologous recombination system of Cre recombinase in the E.coil BJ5183, and naming correct recombinant adenovirus plasmid as pAd-ZBED6; and (3) after Pac I digestion linearization of pAd-ZBED6, transfecting a 293A cell and an original muscle cell of Qinchuan cattle by lipidosomes to obtain recombinant virus particles. The cattle plasmid adenovirus vector pAd-ZBED6 can be applied to functional study and authentication of Qinchuan cattle ZBED6 gene as well as modification of seed cells so as to regulate metabolism of genes related to growth and development of muscle.
Owner:NORTHWEST A & F UNIV
Feed for Qinchuan cattle safely using antibiotics and preparation method thereof
InactiveCN107736480AResidue reductionFast metabolismFood processingAnimal feeding stuffArginineAdditive ingredient
The present invention discloses a feed for Qinchuan cattle safely using antibiotics. The feed mainly comprises accessory materials, an antibacterial ingredient and active ingredients, the antibacterial ingredient is tetracycline, and the active ingredients comprise the following materials in parts by weight: 5-10 parts of malic acids, 2-20 parts of arginine, 10-30 parts of licorice, 3-20 parts ofechinacea purpurea, 8-40 parts of radix ephedrae, 5-30 parts of radix zanthoxyli, and 1-3 parts of compound vitamins; and a weight ratio of the accessory materials, antibacterial ingredient, and active ingredients is 100:2:(2-4.5). A preparation method is as follows: the accessory materials are puffed, then the puffed accessory materials are mixed with the antibacterial ingredient and active ingredients, and the mixture is granulated to prepare a finished product. The feed can significantly reduce tetracycline residue in the Qinchuan cattle body, promotes a weight gain of the cattle, is less in a feeding amount, and has a good palatability; after the accessory materials are puffed, an adsorption capacity of the accessory materials is improved, and a metabolism rate of the antibiotics in the Qinchuan cattle body is further improved. The feed is strongly targeted and only suitable for feeding of the Qinchuan cattle, does not show significant results of other varieties of cattle, and is suitable for wide promotions and applications.
Owner:CHENGDU XIANXIANXIAN BIOTECH CO LTD
Construction and application of a bovine klf3 gene eukaryotic overexpression vector
ActiveCN110777166BExcellent activation abilitySolve problems that affect the expression efficiency of inserted genesMicrobiological testing/measurementGenetically modified cellsBiotechnologyCoding region
The invention discloses the construction and application of a bovine KLF3 gene eukaryotic overexpression vector. Primers were designed for the coding region of Qinchuan cattle KLF3 gene, and the corresponding bovine KLF3 gene was amplified; pcDNA3.1‑KLF3 recombinant overexpression vector was constructed and transfected into cells. By cloning the bovine KLF3 gene and constructing a eukaryotic overexpression vector, the invention can be applied to bovine KLF3 gene function research, identification and regulation of muscle growth and development.
Owner:NORTHWEST A & F UNIV
A method for detecting the cnv marker of Qinchuan cattle gbp2 gene and its application
InactiveCN107119117BSimple and fast operationMicrobiological testing/measurementMarker-assisted selectionPcr ctpp
The invention discloses a method for detecting a CNV (Copy Number Variation) mark of a gene GBP2 of Qinchuan cattle and the application of the CNV mark. The method comprises the following steps: on the basis of a real-time fluorescent quantitation PCR (Polymerase Chain Reaction) technology, by taking the DNA of a whole genome of blood of the Qinchuan cattle as a template, respectively multiplying two CNV regions of the gene GBP2 of the cattle by using two pairs of specific primers GBP2-CNV1 and GBP2-CNV2, multiplying a gene BTF3, which serves as a contrast, of the Qinchuan cattle by using another pair of specific PCR primers R1, and judging whether the copy number of individuals is missing or not according to a calculation result of 2-delta delta Ct. According to the method provided by the invention, on the basis of the relevance between missing of the copy number of the gene GBP2 of the Qinchuan cattle and a growth character, the Qinchuan cattle molecular mark assistant selection breeding work is accelerated favorably. The method is simple, quick and convenient for popularization and application.
Owner:NORTHWEST A & F UNIV
Method for detecting body sizes of Qinchuan cattle through using ANAPC13 gene
ActiveCN102747150BWaist widthEasy to operateMicrobiological testing/measurementMolecular breedingGenotype
The invention discloses a method for detecting the body sizes of Qinchuan cattle through using ANAPC13 gene. The method adopts the combination of a PCR-SSCR technology with DNA sequencing to detect the variation sites of the ANAPC13 gene of the cattle. An SSCP result shows that there are three banding patterns, and a sequencing result shows that there are two variation sites comprising an A-G mutation at 17bp of a first exon and a C-T mutation at 42bp of the first exon. Later sequencing verification shows that the mutations of the two sites in 98% of individuals belong to a linkage mutation. So the three banding patterns are named as AACC, AGCT and GGTT respectively, and the Qinchuan cattle with the number being 404 undergo parting statistics and association analysis of the parting statistics with the body size character. The method is used for auxiliary selection and molecular breeding of the Qinchuan cattle to accelerate the breeding speed of fine breeds of the Qinchuan cattle through screening and detecting molecular genetic markers closely related with the growth and development traits of the Qinchuan cattle in DNA level. The body sizes of the Qinchuan cattle are predicted and determined through the analytic comparison of the genotype, so the method has the advantages of simple operation, low cost, high accuracy, and rapid detection.
Owner:NORTHWEST A & F UNIV
Plasmid type adenovirus vector pAd-NRIP1 and construction method thereof
Construction of a recombinant adenovirus vector of an NRIP1 (nuclear receptor interacting protein 1) gene of Qinchuan cattle can be applied to gene function research and dentification on Qinchuan cattle NRIP1, reconstruction of seed cell, regulation on gene metabolism of muscle, fat and reproduction, etc. The Qinchuan cattle NRIP1 gene is inserted under a CMV promoter of a pAdTrack-CMV adenovirusshuttle plasmid to construct a recombinant shuttle plasmid pAdTrack-CMV-NRIP1. After linearization, a PmeI transforms an E.coli BJ5183 competent cell containing a pAdEasy-1 plasmid; an efficient homologous recombinant system of a Cre recombinase in the E.coli BJ5183 is utilized to complete homologous recombination of pAdTrack-CMV-NRIP1 and pAdEasy-1 in bacteria. A correct recombinant adenovirus plasmid is named as pAd-NRIP1. After digestion linearization of pAd-NRIP1 by a PacI, a liposome is transfected to a 293A cell, so as to obtain a recombinant virosome.
Owner:NORTHWEST A & F UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com