Method for detecting linked mutation of AMPD1 gene of Qinchuan cattle
A technology of gene linkage, Qinchuan cattle, applied in the field of detection of Qinchuan cattle AMPD1 gene linkage mutation, can solve problems such as lack of research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] 1. Collection of experimental animals
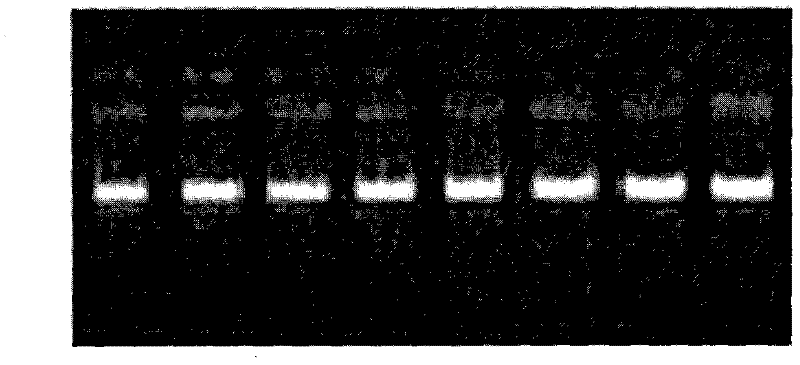
[0066] From September to November 2008, at Qinbao Animal Husbandry Development Co., Ltd. in Shaanxi Province, 30±2-month-old bulls were randomly selected from the Qinchuan cattle population and slaughtered, and a total of 215 Qinchuan cattle blood samples were collected. Add 0.5mL of ACD anticoagulant to 15mL of whole blood, mix upside down and put it in an ice box and bring it back to the laboratory. Store the sample at -80°C for later use.
[0067] 2. Extraction process of genomic DNA from bovine blood samples
[0068] Extraction of genomic DNA from blood samples, the specific operation is as follows:
[0069] (1) After taking out the blood sample from -80°C, put it at room temperature and let it thaw slowly; in the semi-frozen-thawed state, draw 800 μL of blood into a 2 mL sterilized centrifuge tube;
[0070] (2) Draw 800 μL from the PBS buffer to make the total volume reach 1600 μL, and shake gently for 10 min;
[0071] (3)...
Embodiment 2
[0106] 1. Design of PCR primers for the 11th intron of Qinchuan cattle AMPD1 gene
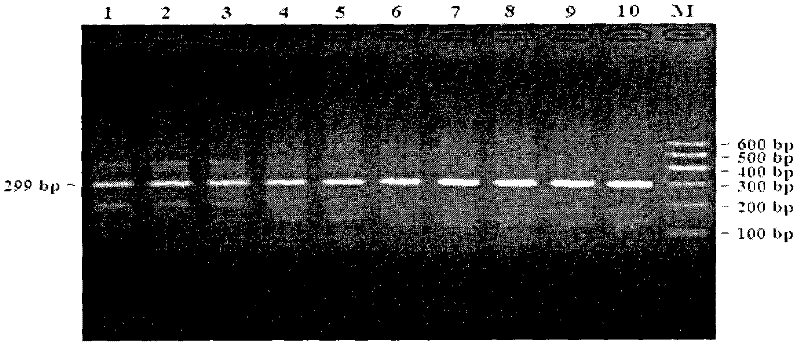
[0107] Taking the bovine (accession number: NC_007301) sequence published by GenBank in the NCBI database as a reference, the PCR primers (fragment size: 299bp) of the 11th intron of the AMPD1 gene of Qinchuan cattle were designed using Premier 5.0 software combined with Oligo 6.0 software. The primer sequence as follows:
[0108] Upstream primers:
[0109] 5'-AACCCTCAGGCTCACCCA-3'18bp;
[0110] Downstream primers:
[0111] 5'-GGGCTTAGGGCTCTTGGA-3' 18bp.
[0112] The primer pair amplifies the 11th intron region of Qinchuan cattle AMPD1 gene.
[0113] 2. PCR amplification
[0114] (1) PCR reaction system
[0115] The PCR reaction system adopts the mixed sample addition method. In the experiment, we usually calculate the amount required for each component first, then calculate the total amount of sample addition, and then divide it into each PCR tube equally, then add template DNA, and the...
Embodiment 3
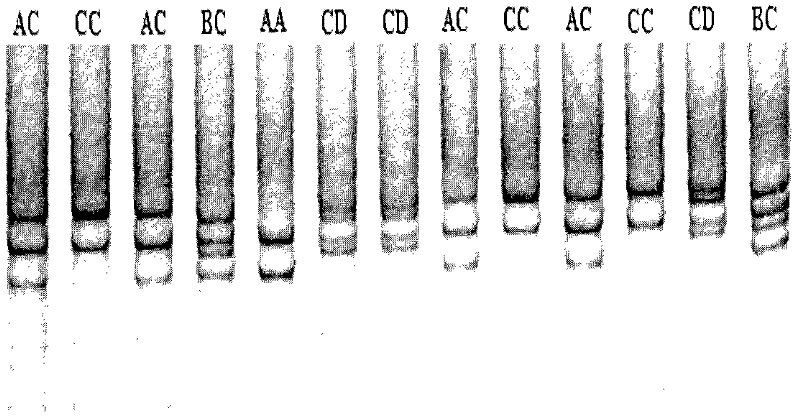
[0137] According to the electropherogram results after sequencing of individual and mixed DNA templates, using the bovine NC_007301 sequence published by GenBank in the NCBI database as a reference, the sequenced sequence and the published reference sequence were sequenced using BioXM (Version 2.6) software and DNA Star 7.10 software. Comparison and mutation site analysis showed that the base mutation site was quickly screened. In order to further verify the variation of the base mutation site, the next experiment was carried out in the entire Qinchuan cattle population.
[0138] Example 4: Polyacrylamide gel electrophoresis analysis of Qinchuan cattle AMPD1 gene
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com