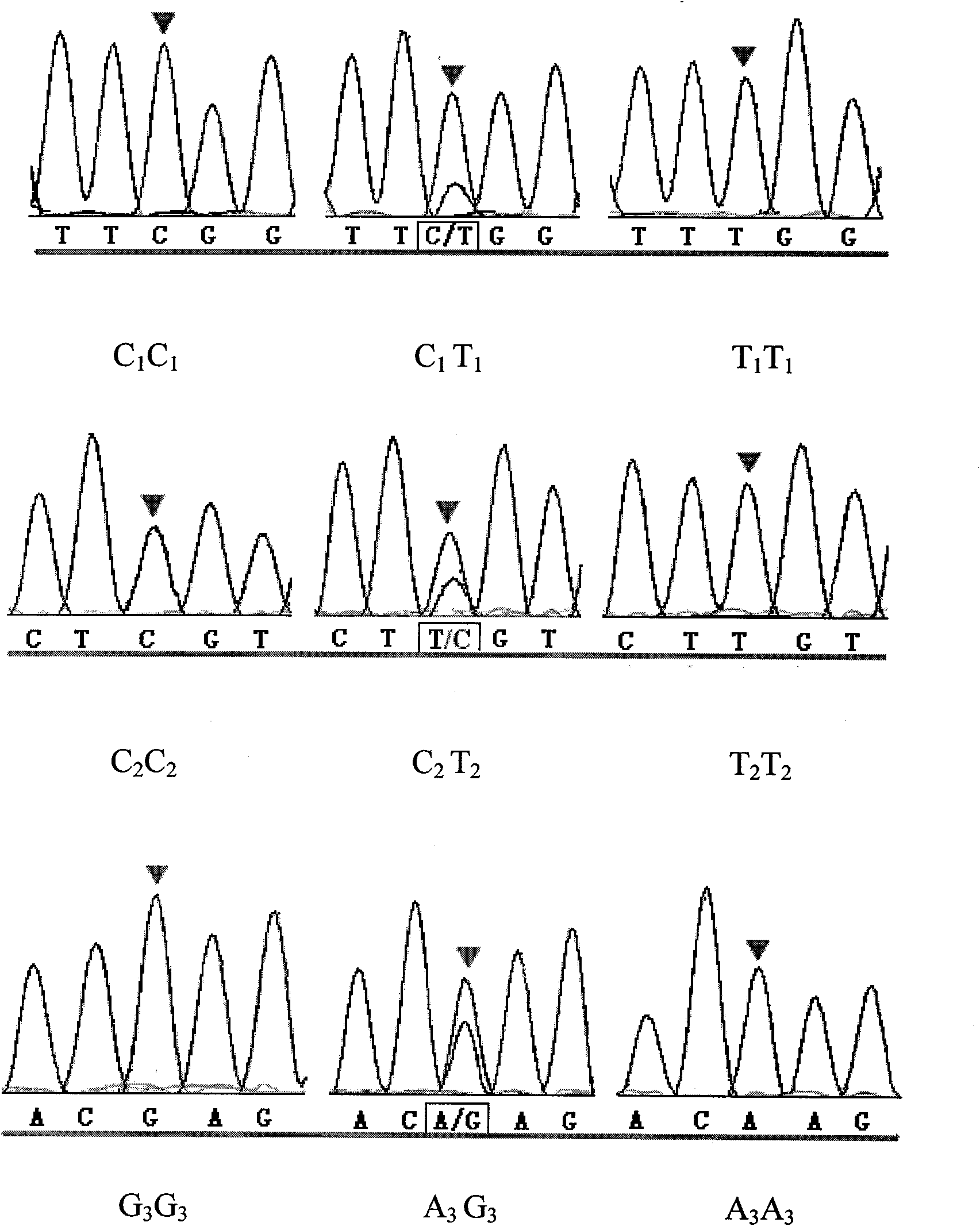
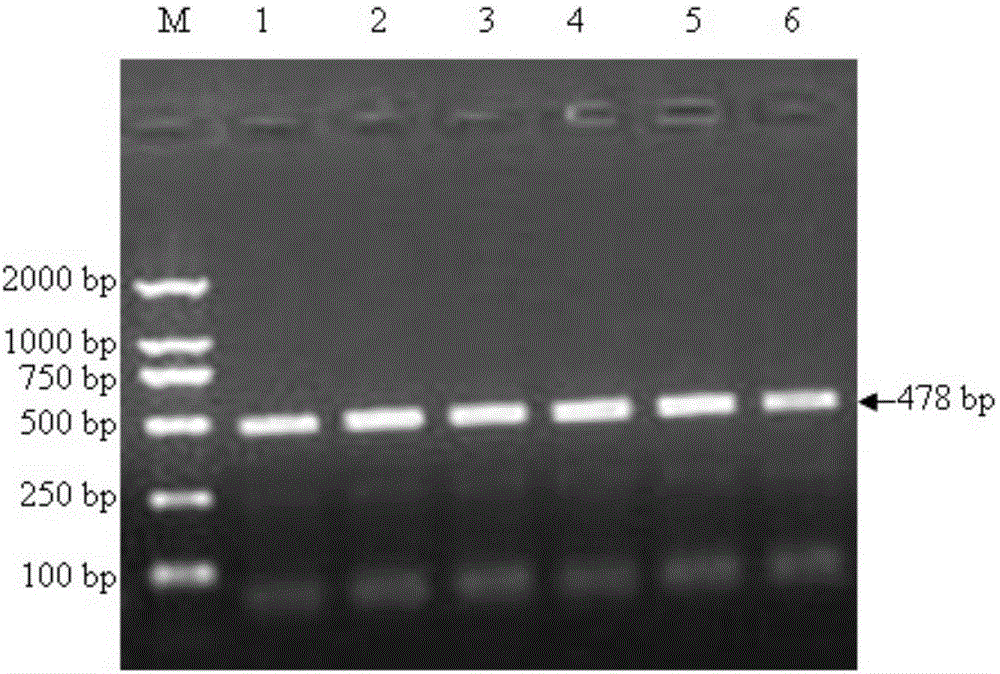
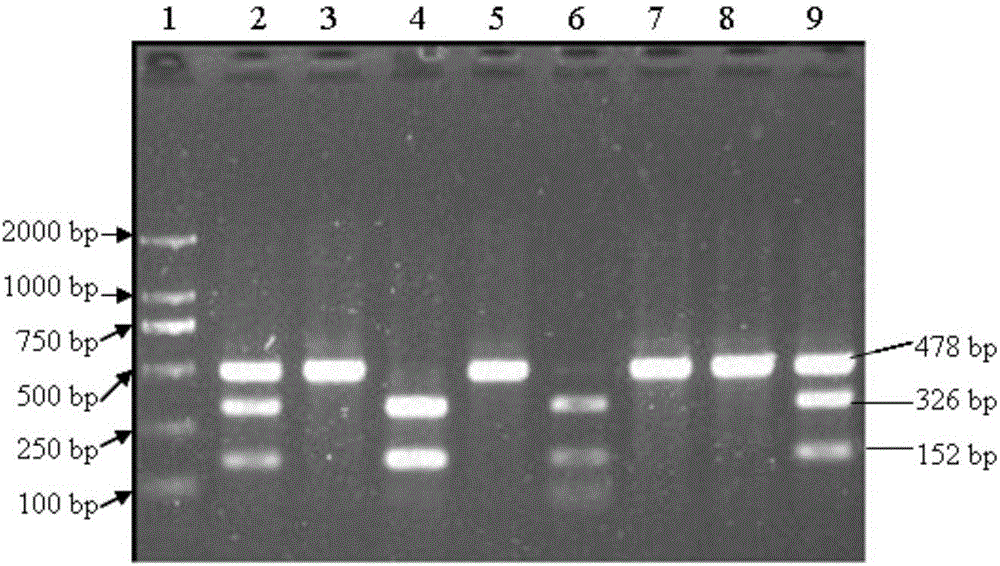
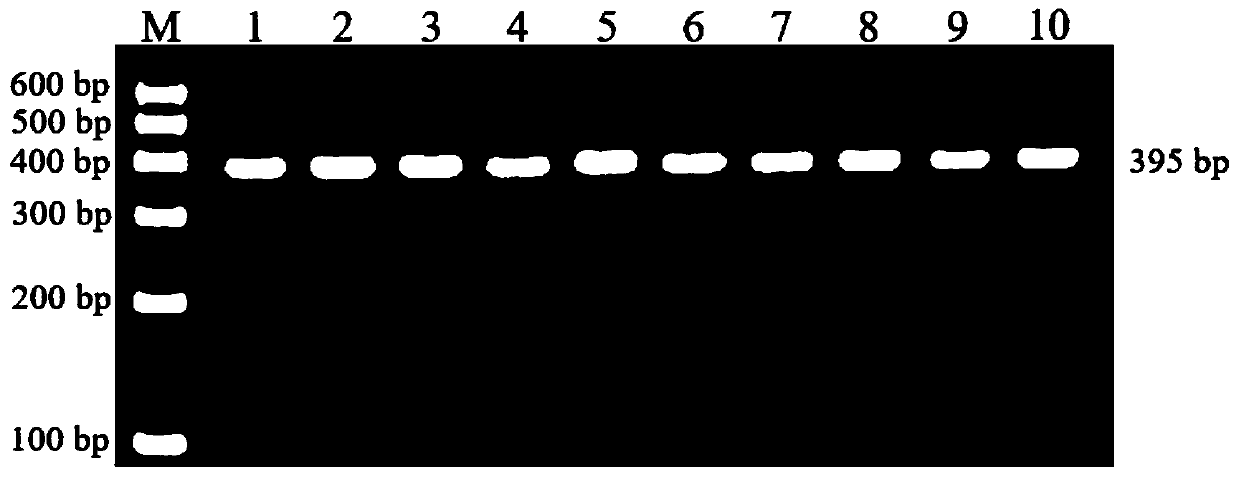
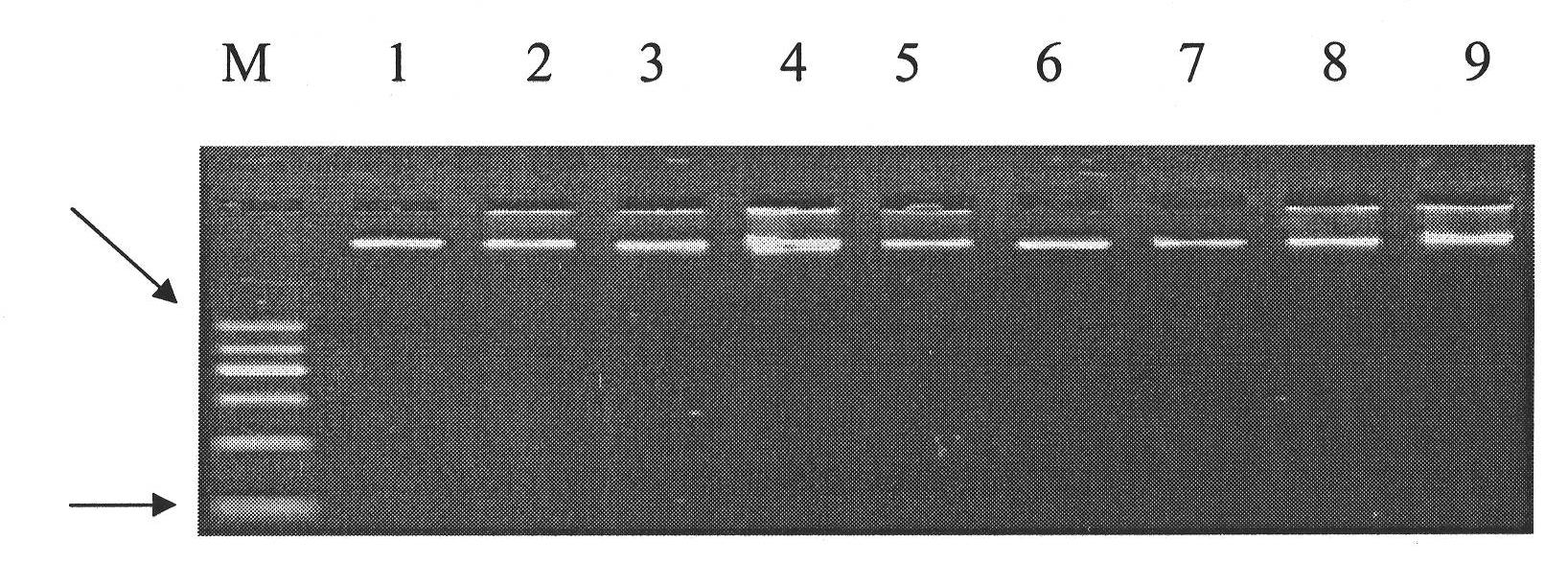
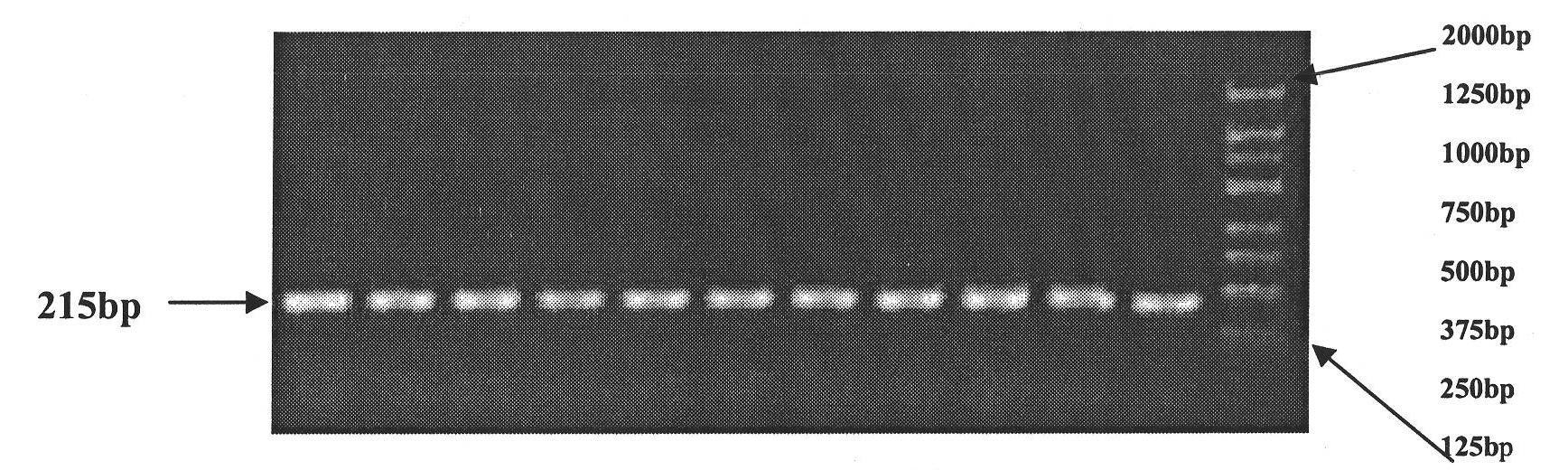
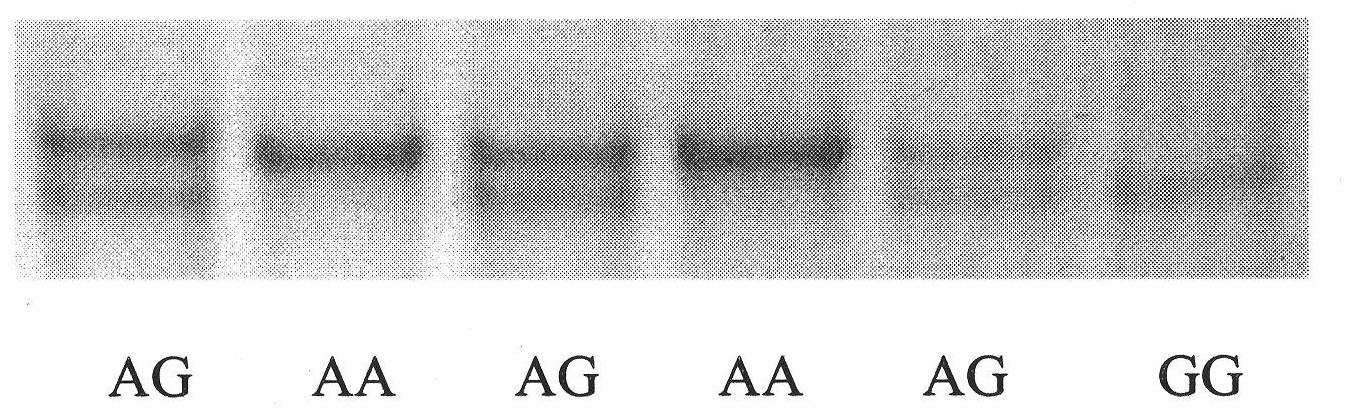
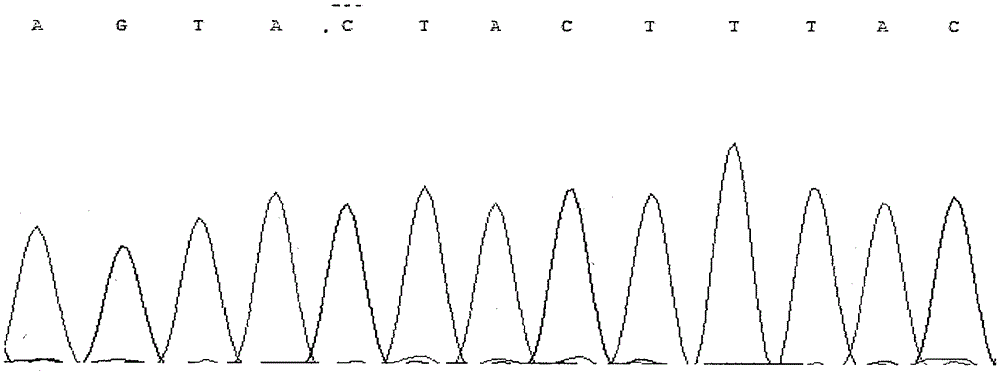
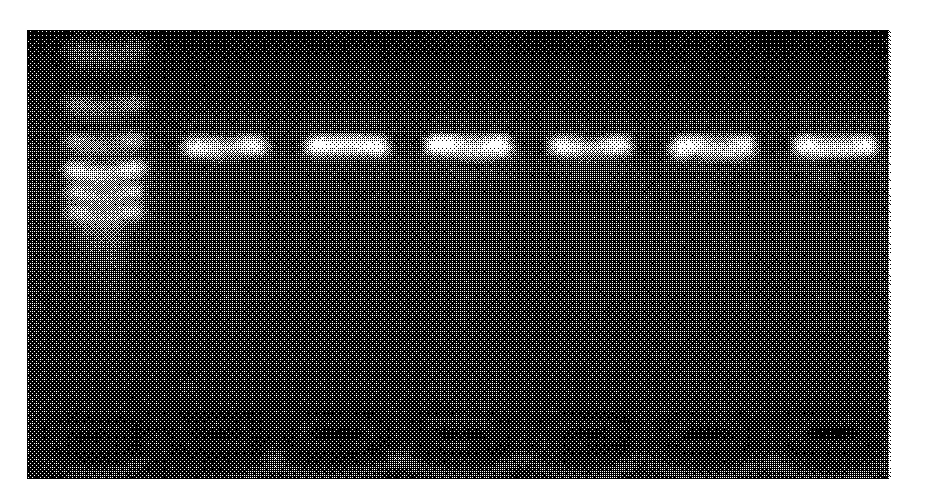Patents
Literature
43results about How to "Accurately estimate breeding values" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Single nucleotide polymorphism of SCD genes in dairy goat and detection method thereof
InactiveCN101705290AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementElectrophoresisMolecular breeding
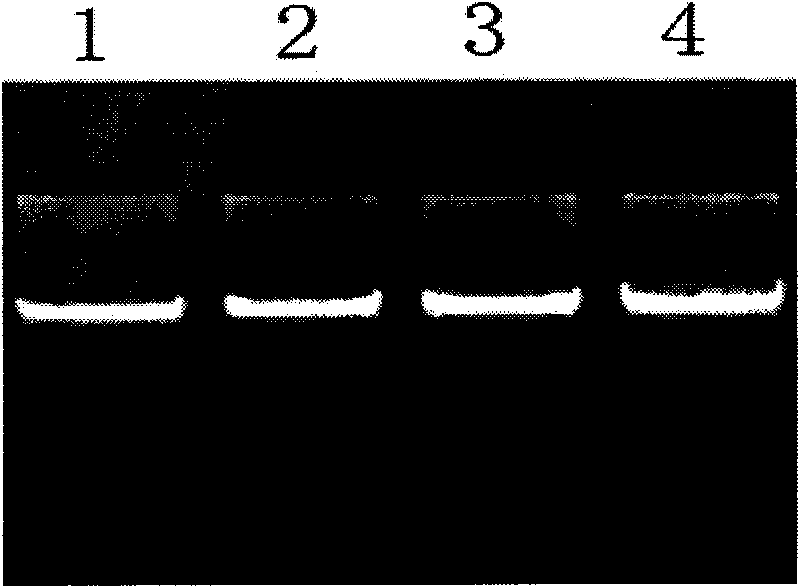
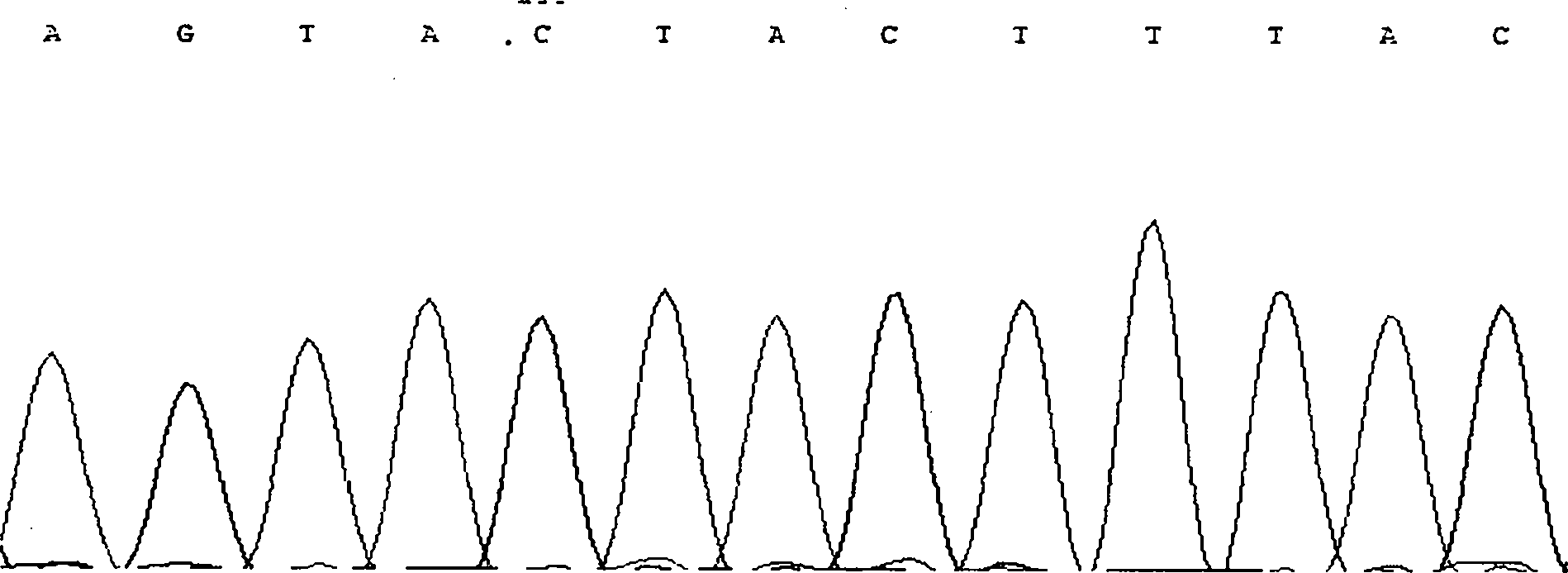
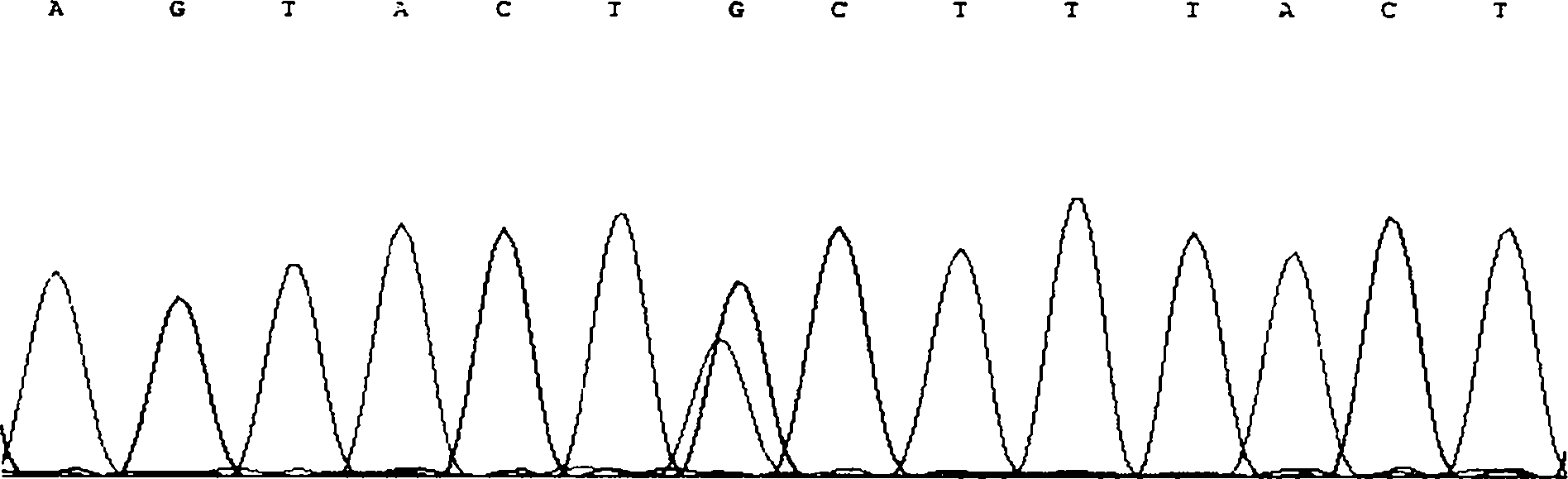
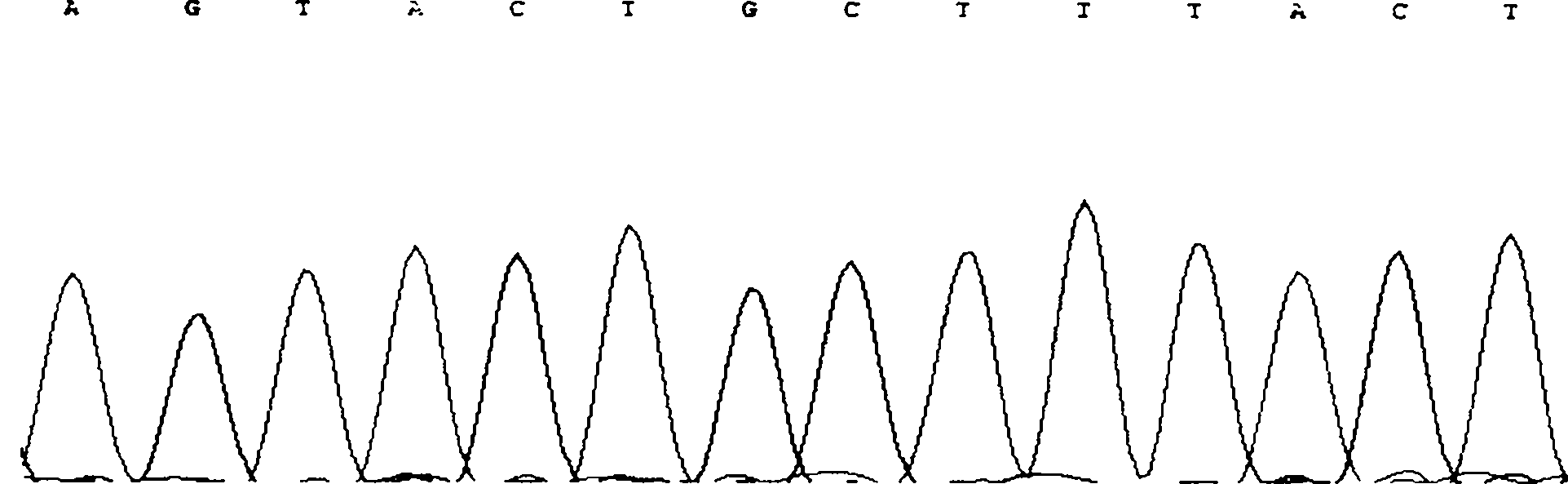
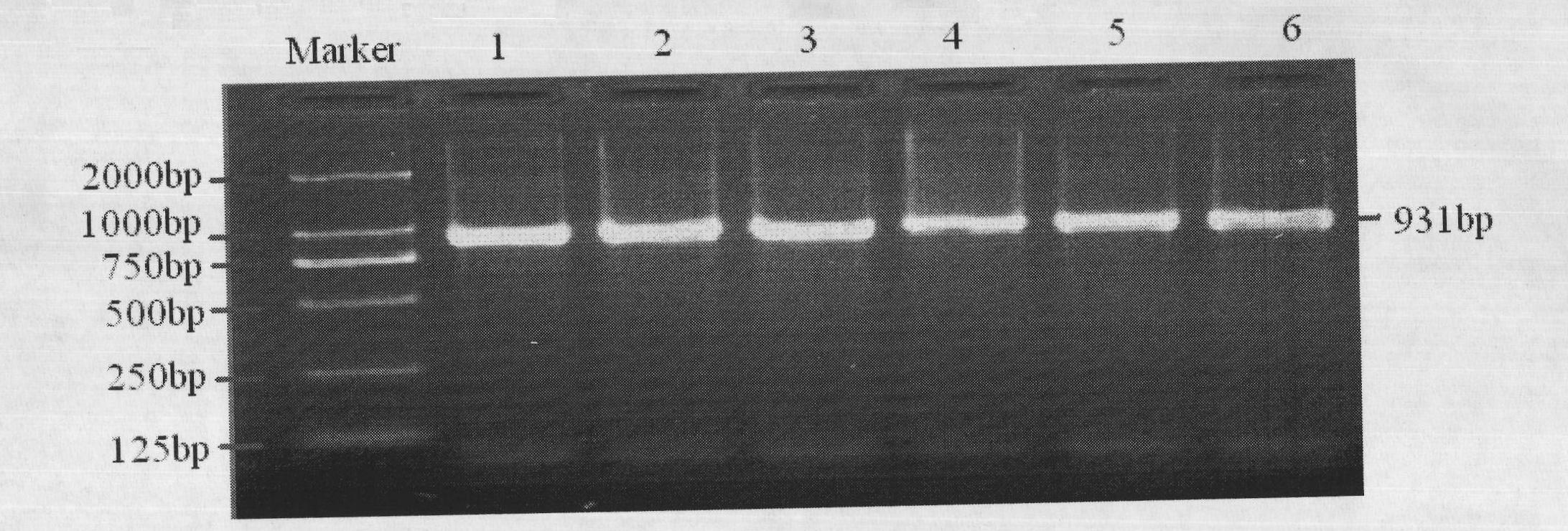
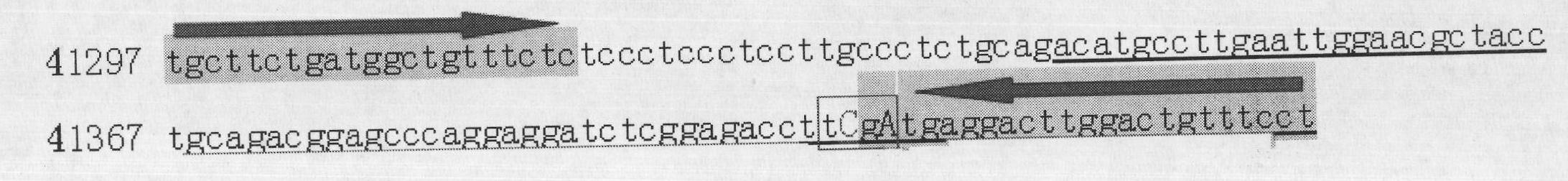
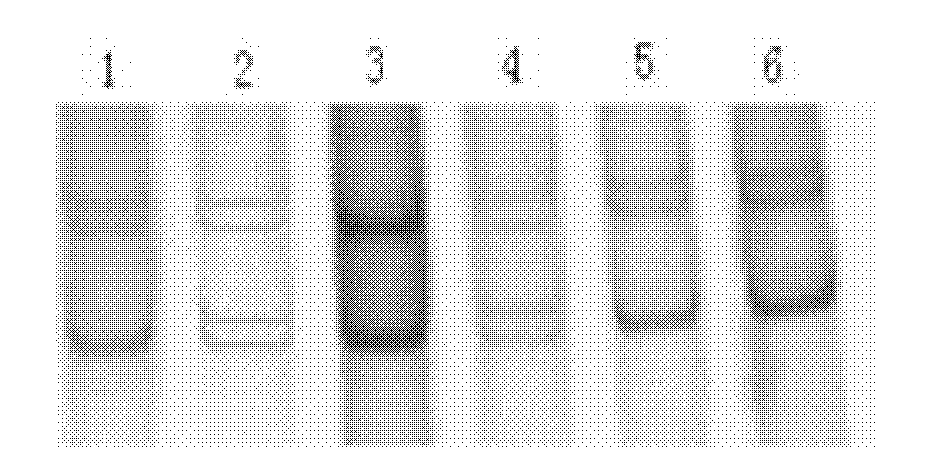
The invention discloses single nucleotide polymorphism of SCD genes in dairy goat and a detection method thereof. The single nucleotide polymorphism of the genes comprises G / A single nucleotide polymorphism which is the 5001st in the SCD genes in the dairy goat. The detection method comprises the following steps: taking the DNA of the whole genome of the dairy goat to be detected containing the SCD genes as a template and a primer pair P as a primer to carry out PCR amplification on the SCD genes in the dairy goat; after using restriction enzyme NdeI to digest the PCR amplified product, carrying out agarose gel electrophoresis on amplified fragments after restriction enzyme; and identifying the 5001bp base polymorphism of the SCD genes in the dairy goat according to the electrophoresis results. The method screens and detects the molecular genetic markers closely related to the milk production traits of the dairy goat on the DNA level to be used for assisted selection and molecular breeding of the dairy goat and speed up find breeding of the dairy goat.
Owner:NORTHWEST A & F UNIV
Method for breeding kidding traits by utilizing 3-gene pyramiding effect
InactiveCN103525921AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementInteinA-DNA
The invention discloses a method for breeding kidding traits by utilizing 3-gene pyramiding effect. The method comprises the following steps: amplifying a 3' untranslated region of a stem cell factor (SCF) gene, an exon 7 of a III type transmembrane tyrosine kinase receptor (KIT) gene and an intron 1 of a kiss (KISS1) gene respectively by using three pairs of primers and by taking goat genome DNA as a template; judging the size of each amplification product by using agarose gel electrophoresis at the concentration of 1.5%; screening the site mutation of the amplification products of the three pairs of primers by a DNA sequencing technology; performing genetic typing and gene frequency analysis on the single nucleotide polymorphisms (SNPs) of three sites of the SCF gene, the KIT gene and the KISS1 gene by using polyacrylamide gel electrophoresis at the concentration of 10% and agarose gel electrophoresis at the concentration of 3.5%; analyzing the relationship between different genotype combinations and kidding quantity; and screening out the optimal genotype combination.
Owner:NORTHWEST A & F UNIV
Qinchuan cattle FoxO1 gene mononucleotide polymorphism molecular marker detection method and application
InactiveCN103233001AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationNucleotideElectrophoresis
The invention discloses cattle FoxO1 gene mononucleotide polymorphism and a detection method thereof. The gene mononucleotide polymorphism comprises that: with a Qinchuan cattle whole genome DNA comprising the FoxO1 gene as a template, and with a primer pair P as primers, the Qinchuan cattle FoxO1 gene is subjected to PCR amplification; a restriction endonuclease HhaI is used for digesting the PCR amplification product, and the digested segment is subjected to agarose gel electrophoresis; and according to the electrophoresis result, base polymorphism on the 178132 site of the Qinchuan cattle FoxO1 gene is identified. The FoxO1 gene is important for animal muscle fat traits and growth traits. With the method, the molecular genetic marker closely related to Qinchuan cattle growth traits is screened and detected on a DNA level. The method is used in Qinchuan cattle assisted selection and molecular breeding, and assists in accelerating Qinchuan cattle breeding speed.
Owner:NORTHWEST A & F UNIV
Multi-gene pyramiding breeding method for thoroughbred milk goats
InactiveCN102605064AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementExonProlactin receptor
The invention discloses a multi-gene pyramiding breeding method for thoroughbred milk goats. Genome DNA (deoxyribonucleic acid) of a milk goat continuously giving birth to two or more lambs is used as a template, four pairs of primers respectively expand intron 2 and exon 10 of a prolactin receptor gene and an untranslation region (5'UTR) and exon 1 of luteotropin beta calcmeurin 5', the sizes of expanded products are judged by agarose gel electrophoresis, site mutation of the expanded products of the four pairs of primers are screened by DNA sequencing technology, then polyacrylamide gel electrophoresis is used for performing genetic typing and gene frequency analysis for SNPs (single nucleotide polymorphisms) of four sites of the prolactin receptor gene and the luteotropin beta calcmeurin, the relation of polymorphism of a multiple-birth milk goat individual (F1 generation) and the number of born lamps and the relation of different genetype combinations and the number of the born lamps are analyzed, parental generation (F0 generation) is reviewed, filial generation (F2 generation) is tracked, the relation of the genetype combination of an ewe individual and the number of the born lamps is detected, and contribution of different genetypes in terms of prolific trait formation is analyzed.
Owner:NORTHWEST A & F UNIV
Detection method for single nucleotide polymorphism of STMN1 gene of Beijing duck and molecular markers thereof
InactiveCN102816759AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementAnimal husbandryElectrophoresisExon
The invention discloses single nucleotide polymorphism of the STMN1 gene of a Beijing duck and a detection method thereof. The detection method for the single nucleotide polymorphism of the gene comprises the following steps: with whole genome DNA of a to-be-detected Beijing duck containing the STMN1 gene as a template and the primer pair P as primers, carrying out PCR amplification on the STMN1 gene of the Beijing duck; digesting a PCR amplification product with restriction endonuclease and then carrying out agarose gel electrophoresis on amplified segment after digestion; and identifying polymorphism of the 48th nucleotide base of exon 4 of the STMN1 gene of the Beijing duck according to results of electrophoresis. According to the invention, molecular genetic markers closely related to economic characters of the Beijing duck are screened and detected at the level of DNA, which provides molecular bases for rapid selective breeding of the Beijing duck.
Owner:XUZHOU NORMAL UNIVERSITY
Method for selecting molecular marker for goat yeaning traits
InactiveCN101899526AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementBiotechnologyPolymerase L
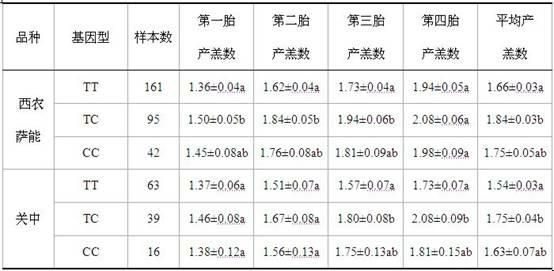
The invention discloses a method for selecting a molecular marker for goat yeaning traits, which comprises the following steps of: taking a goat genome DNA sequence as a template, amplifying KITL gene introns 1 and 6 by using primers P1 and P2 under a PCR condition in the presence of TaqDNA polymerase, buffer environment, Mg2+ and dNTPs respectively, and judging the size of a destination fragment according to an agarose gel electrophoresis result; digesting the PCR amplification product of the primer P1 by using restriction enzyme CviAII, and then detecting the enzyme digested amplification fragment by using polyacrylamide gel electrophoresis, wherein the amplification product of the primer P1 has mutation of two basic groups; detecting the PCR amplification product of the primer P2 by adopting the polyacrylamide gel electrophoresis, wherein the amplification product of the primer P2 has mutation of one basic group; and then performing gene analysis and gene frequency analysis on the amplification products of the primers P1 and P2, and performing association analysis between the amplification products and the yeaning numbers of Guanzhong dairy goats, western Saanen dairy goats and Boer goats, wherein the analysis results show that an SNPs site of the KITL gene, detected by the primers P1 and P2, can be taken as the molecular marker for goat yeaning trait selection.
Owner:NORTHWEST A & F UNIV
Method for detecting single nucleotide polymorphism of sheep KITLG gene and application of method
ActiveCN106498078AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementKITLG GeneLitter
The invention discloses a method for detecting single nucleotide polymorphism of a sheep KITLG gene and application of the method. The method comprises the following steps: carrying out PCR amplification on fragments of the sheep KITLG gene in a manner of taking a whole-genome DNA of sheep to be detected as a template and taking a primer pair P as primers; digesting a PCR amplification product with restriction endonuclease SspI, and then, carrying out agarose gel electrophoresis on the amplified fragments subjected to digestion; identifying the polymorphism of a 47569-th base of the sheep KITLG gene according to electrophoresis results. The mutant site of the base is closely associated with reproduction traits, i.e., litter size of the sheep, so that the method is a method for detecting molecular genetic markers, which are closely related to the reproduction traits of the sheep, from a DNA level, and can be applied to the assisted selection and molecular breeding of the sheep, and thus, the good-variety breeding speed of the sheep is accelerated.
Owner:天津奥群羊业研究院有限公司
Molecular marking method of using neuroendocrine factor genes to select kidding characters
InactiveCN101906480AImprove selection efficiencySpeed up breeding progressMicrobiological testing/measurementSECRETOR FACTORGenomic DNA
The invention discloses a molecular marking method of using neuroendocrine factor genes to select kidding characters. The method comprises the following steps of: using a goat genomic DNA sequence as a template; amplifying Kisspeptin gene intron 2 by using a primer P1 under the PCR condition in the presence of TaqDNA polymerase, buffering environment, Mg2+, dNTPs, and then judging the size of the target fragment by agarose gel electrophoresis; detecting the PCR application product of the primer P1 by polyacrylamide gel electrophoresis, finding that the amplification SNPs of the primer P1 has one-base mutation and 8bp-base deletion, and then carrying out genotyping and gene frequency analysis on the amplification product of the primer P1, and the correlation analysis with the kidding characters of Guanzhong diary goat and Xinong Saanen dairy goat. Shown by experiments, the SNPs of the Kisspeptin gene intron 2 detected by the primer P1can be used as molecular marker of the selection of kidding characters.
Owner:NORTHWEST A & F UNIV
Single nucleotide polymorphism of GHRHR genes in dairy goat and detection method thereof
InactiveCN101705289AAccurate estimateImprove selection efficiencyMicrobiological testing/measurementEnzymeMolecular breeding
The invention discloses single nucleotide polymorphism of GHRHR genes in dairy goat and a detection method thereof. The single nucleotide polymorphism of the genes comprises C / G base polymorphism which is the 231st in the sequence table of the GHRHR gene in the dairy goat shown in SEQ ID NO.1. The detection method comprises the following steps: taking the DNA of the whole genome of the dairy goat to be detected containing the GHRHR genes as a template and a primer pair P as a primer to carry out PCR amplification on the GHRHR genes in the dairy goat; after using restriction enzyme MspI to digest the PCR amplified product, carrying out agarose gel electrophoresis on amplified fragments after restriction enzyme; and identifying the polymorphism of the genes according to the electrophoresis results. The method screens and detects the molecular genetic markers closely related to the milk production traits of the dairy goat on the DNA level to be used for assisted selection and molecular breeding of the dairy goat and speed up find breeding of the dairy goat.
Owner:NORTHWEST A & F UNIV
Method for selecting milk production characters to breed milk goat by utilizing double gene polymerization effect
InactiveCN103468819AImprove selection efficiencySpeed up breeding progressMicrobiological testing/measurementAnimal husbandryProlactin receptorExon
The invention discloses a method for selecting milk production characters to breed a milk goat by utilizing the double gene polymerization effect. Milk goat genome DNA serves as a template, two pairs of primers are used for amplifying the exon 9 of a gene of a PRLR and the intron 1 of a gene of an ELF5 respectively, agarose gel electrophoresis with the concentration of 1.5% is used for judging the sizes of the amplified products, the locus mutations of the amplified products of the two pairs of primers are screened based on the DNA sequencing technique, then, the agarose gel electrophoresis with the concentration of 3.5% is used for carrying out genetic typing and gene frequency analyzing on SNPs of the two locuses of the gene of the PRLR and the gene of the ELF5, the relation between different gene type combinations and the milk production characters is analyzed, and the optimal gene combination is screened out.
Owner:NORTHWEST A & F UNIV
Method for detecting single nucleotide polymorphism of sheep PCNP (PEST-Containing Nuclear Protein) gene by using PCR-RFLP (Polymerase Chain Reaction-Restriction Fragment Length Polymorphism) and application of method
ActiveCN106755371AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementMolecular geneticsBiology
The invention discloses a method for detecting single nucleotide polymorphism of a sheep PCNP (PEST-Containing Nuclear Protein) gene by using PCR-RFLP (Polymerase Chain Reaction-Restriction Fragment Length Polymorphism) and application of the method. The method comprises the following steps: carrying out PCR amplification on a sheep PCNP gene segment by taking a sheep whole genome DNA to be measured as a template and taking a primer pair P as a primer; after a PCR amplification product is digested by a restriction enzyme SPhI, carrying out agarose gel electrophoresis on the digested amplified segment; authenticating the single nucleotide polymorphism of the 5019th site of the sheep PCNP gene according to an electrophoresis result. As a base mutation site is closely associated with the kidding number of sheep reproduction traits, the method is a method for detecting molecular genetic markers closely associated with the sheep reproduction traits at the DNA level, and can be used for auxiliary selection and molecular breeding of sheep and increasing the improved variety breeding speed of the sheep.
Owner:甘肃润牧生物工程有限责任公司
Method for detecting single nucleotide polymorphism of cattle krupple-like factor (KLF) 7 gene
InactiveCN101899500AExcellent genetic resourcesAccurately estimate breeding valuesMicrobiological testing/measurementEnzyme digestionNucleotide
The invention discloses a method for fast detecting the single nucleotide polymorphism (SNP) of a cattle krupple-like factor (KLF) 7 gene. The method comprises the following steps of: performing polymerase chain reaction (PCR) amplification on the cattle KLF7 gene by taking a whole genome DNA comprising a KLF7 gene of cattle to be detected as a template and the mixture of primer pairs P1, P2, P3 and the like in a mol ratio as a primer; after digesting a PCR amplification product by using a restriction enzyme TaqI, performing agarose gel electrophoresis on an amplified fragment subjected to enzyme digestion; and according to the result of the agarose gel electrophoresis, identifying the SNP of the 41041st, 42025th and 42075th positions of the cattle KLF7 gene. In the method, the SNPs of the KLF7 gene are subjected to genotyping and gene frequency analysis, and the growth traits of Nanyang cattle in different growth stages are subjected to trait association analysis. The results show that the SNP locus for the KLF7 gene, particularly P2, can serve as a molecular marker selected in the early stage (6 month age and 12 month age) of the growth traits of the cattle.
Owner:NORTHWEST A & F UNIV
Method for detecting base mutation polymorphism of goat gonadotropin releasing hormone and growth differentiation factor 9
InactiveCN101845507AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementMaterial analysis by electric/magnetic meansBiotechnologyIntein
The invention discloses a method for detecting the polymorphism of three base mutations of the gene exon 4 of goat gonadotropin releasing hormone and the gene exon 2 of the growth differentiation factor 9. The method comprises the following steps: using the DNA sequence of goat genome as template, using 2 to amplify the primer P1 and P2 under PCR condition in the presence of Taq DNA polymerase, buffer environment, Mg2+ and dNTPs, using agarose gel electrophoresis to judge the size of the target fragments; using polyacrylamide gel electrophoresis to detect the PCR amplified product of the primer P1 and P2 to find that the two amplification sites have three base mutations, then performing genotyping and gene frequency analysis to the SNPs of the two genes, and performing association analysis with the fecundity traits of Boer goat and Xinong Saanen dairy goat. The result shows that the SNPs sites detected by the primer P1 and P2, of the GnRH gene exon 4 (containing partial intron 3 and 3' non-translational region) and the GDF9 gene exon 2, thus the method can be used for molecular marking of fecundity trait selection of goat.
Owner:NORTHWEST A & F UNIV
Method for detecting insertion-deletions polymorphism of sheep FTH-1 (ferritin heavy polypeptide1) genes by utilizing PCR-SSCP (polymerase chain reaction-single strand conformation polymorphism) and application thereof
ActiveCN106701930AExcellent genetic resourcesAccurately estimate breeding valuesMicrobiological testing/measurementInsertion deletionSingle-strand conformation polymorphism
The invention discloses a method for detecting insertion-deletions polymorphism of sheep FTH-1 (ferritin heavy polypeptide1) genes by utilizing PCR-SSCP (polymerase chain reaction-single strand conformation polymorphism) and application thereof. The method comprises the following steps: taking to-be-detected sheep whole genome DNA containing FTH-1 genes as a template, performing PCR amplification on the sheep FTH-1 gene segments, and performing detection and genotyping on the PCR product by utilizing the SSCP technology; and identifying the 639th locus insertion-deletions polymorphism of the sheep FTH-1 genes according to electrophoretic results. The method is a method for screening and detecting a molecular genetic marker which is closely related to reproductive performances of sheep on the DNA level so as to be used for assistant selection and molecular breeding of the sheep and acceleration of sheep stock breeding.
Owner:甘肃润牧生物工程有限责任公司
Method for detecting single nucleotide polymorphism of sheep FTH-1 (Ferritin Heavy Polypeptide-1) gene by using PCR-RFLP (Polymerase Chain Reaction-Restriction Fragment Length Polymorphism) and application of method
ActiveCN106755370AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementMolecular geneticsPolymorphism Detection
The invention discloses a method for detecting single nucleotide polymorphism of a sheep FTH-1 (Ferritin Heavy Polypeptide-1) gene by using PCR-RFLP (Polymerase Chain Reaction-Restriction Fragment Length Polymorphism) and application of the method. The method comprises the following steps: carrying out PCR amplification on a sheep FTH-1 gene segment by taking a sheep whole genome DNA to be measured as a template and taking a primer pair P as a primer; after a PCR amplification product is digested by a restriction enzyme XspI, carrying out agarose gel electrophoresis on the digested amplified segment; authenticating the single nucleotide polymorphism of the sheep FTH-1 gene according to an electrophoresis result. As the polymorphism of a base mutation site is closely associated with the kidding number of sheep reproduction traits, the polymorphism detection method is a method for detecting molecular genetic markers closely associated with the sheep reproduction traits at the DNA level, and can be used for auxiliary selection and molecular breeding of sheep and increasing the improved variety breeding speed of the sheep.
Owner:甘肃润牧生物工程有限责任公司
SNP molecular marker for CFL1 gene of Qinchuan cattle and detection method of SNP molecular marker
ActiveCN110564867AExcellent genetic resourcesAccurately estimate breeding valuesMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionElectrophoresis
The invention belongs to the field of molecular genetics, and provides an SNP molecular marker for CF1 gene of Qinchuan cattle and a detection method of the SNP molecular marker. The whole genome DNAof Qinchuan cattle to be tested containing the CFL1 gene is used as a template, a primer pair P is used as primers, and PCR is carried out to amplify the CFL1 gene; agarose gel electrophoresis is performed on the amplified fragments after enzyme digestion, after the PCR amplified product is digested with a restriction enzyme HinfI; and the base polymorphism at position 2052 of the CFL1 gene of Qinchuan cattle is identified according to the electrophoresis results. Because the mutation site is closely related to the beef traits (body length, chest width, chest depth and weight) of Qinchuan cattle, and the method is a method for screening and detecting the molecular genetic markers closely related to the growth traits of Qinchuan cattle at DNA level, the SNP molecular marker and the method can be used in auxiliary selection and molecular breeding of Qinchuan cattle, so as to accelerate the breeding speed of Qinchuan cattle.
Owner:YANGZHOU UNIV
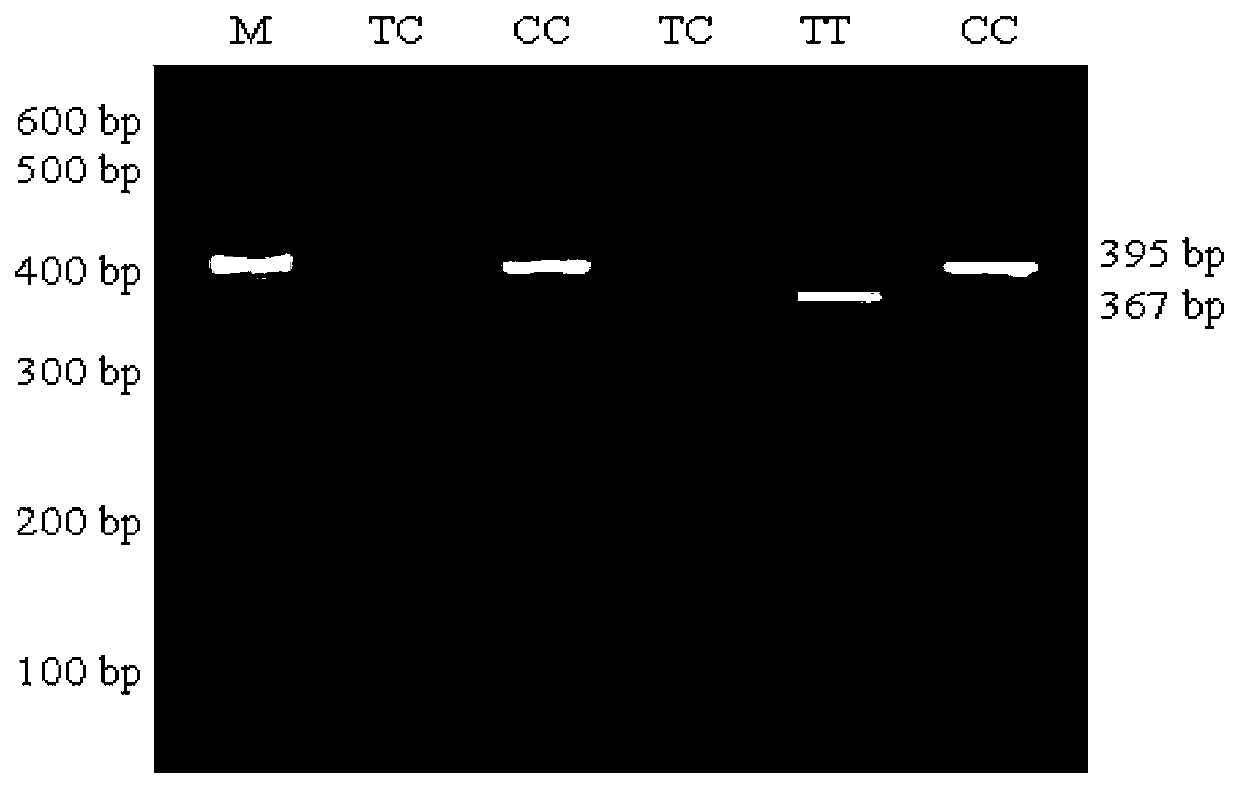
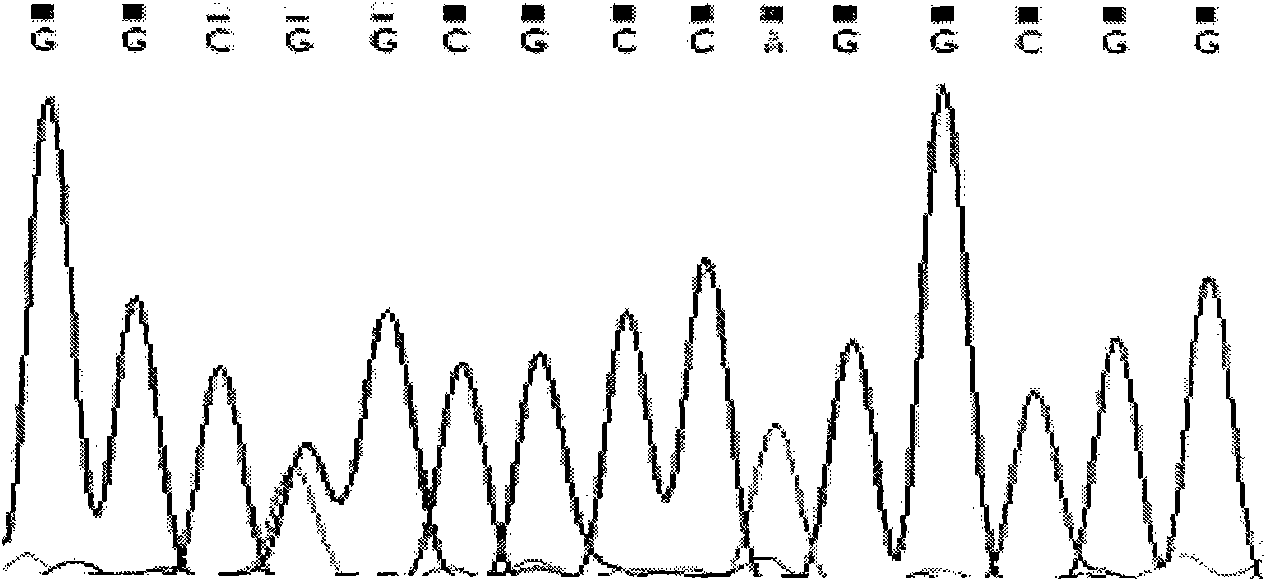
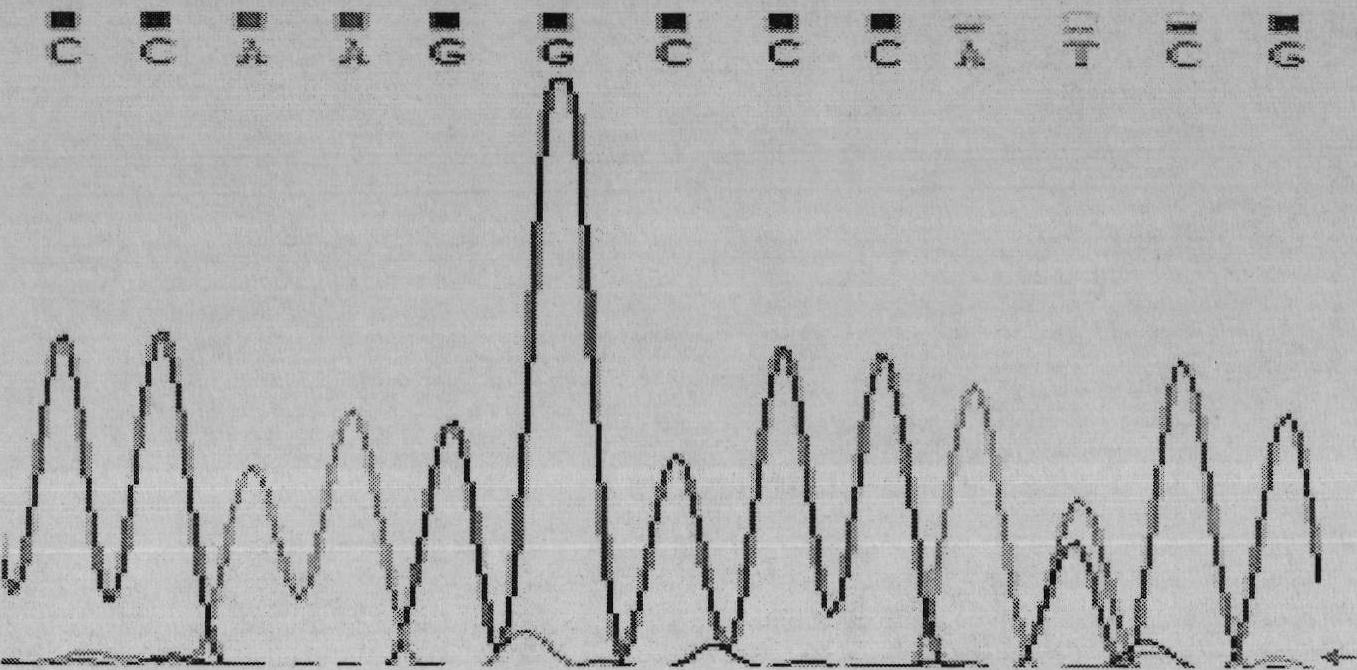
Single nucleotide polymorphic locus of cattle I-mfa gene and detection method thereof
InactiveCN102660540AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationNucleotideElectrophoresis
The invention discloses a single nucleotide polymorphic locus of a cattle I-mfa gene and a detection method thereof. The detection method of the single nucleotide polymorphisms locus of cattle I-mfa gene comprises utilizing cattle whole-genome deoxyribonucleic acid (DNA) to be detected containing the I-mfa gene as a template, utilizing a primer pair P as primers, performing polymerase chain reaction (PCR) amplification on the cattle I-mfa gene and performing polyacrylamide gel electrophoresis on amplified framorphism; according to electrophoresis results, identifying the 12284 locus of the cattle I-mfa gene to be nucleotide polymplate of A or G; and identifying the 12331 locus of the cattle I-mfa gene to be nucleotide polymorphism of T or C. The two locus is completely in linkage. The detection method of the single nucleotide polymorphic locus of the cattle I-mfa gene is a method which screens and detects molecular genetic markers closely related to growth traits of cattle from the DNA level, and provides the molecular basis for cattle crossbreeding utilizing the crossbreed advantages.
Owner:XUZHOU NORMAL UNIVERSITY
Meat-duck OTXR gene single nucleotide polymorphism and detecting method and application thereof
ActiveCN105603099AAccurate estimateImprove selection efficiencyMicrobiological testing/measurementEnzymeDigestion
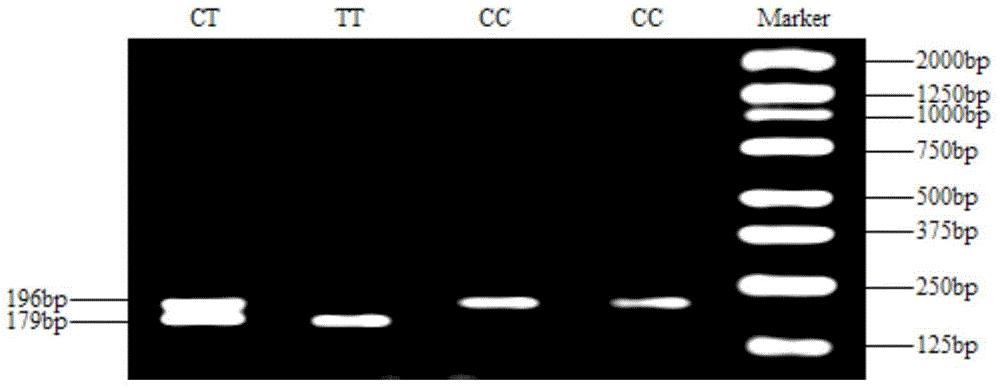
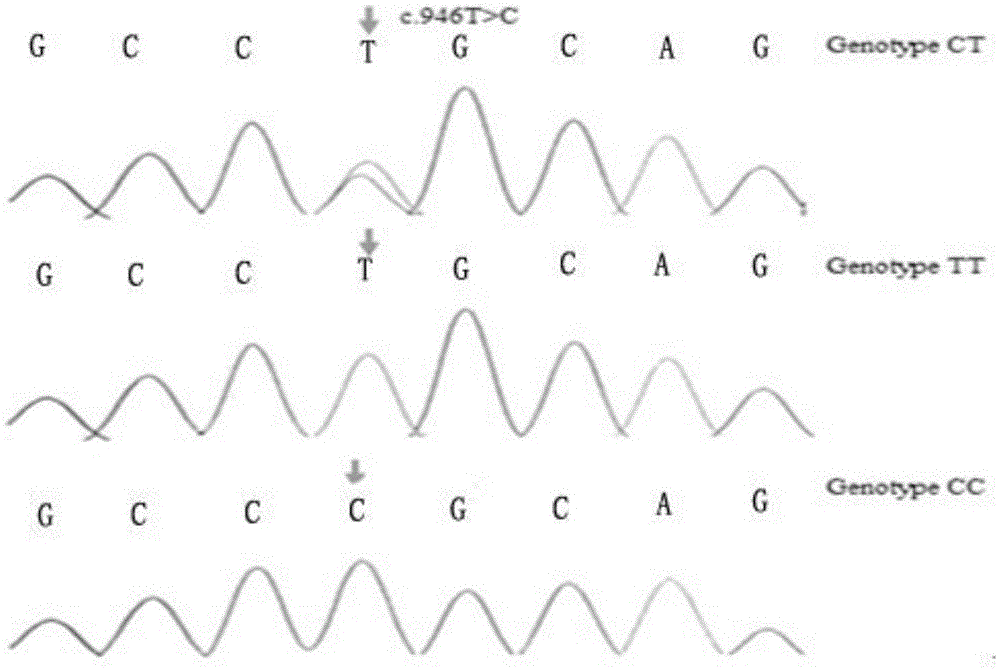
The invention discloses meat-duck OTXR gene single nucleotide polymorphism and a detecting method and application thereof, and belongs to the field of molecular genetics detecting. The detecting process of the gene single nucleotide polymorphism includes the steps that to-be-detected meat-duct whole genome DNA containing OTXR genes serves as a template, a primer pair P serves as a primer, and the meat-duck OTXR genes are subjected to PCR amplification; after the PCR amplification product is digested through restriction enzymes PstI, amplified fragments obtained after digestion are subjected to agarose gel electrophoresis; the polymorphism of basic groups of the 946th nucleotide of second exons of the meat-duck OTXR genes is identified according to the electrophoresis result. According to the meat-duck OTXR gene single nucleotide polymorphism and the detecting method and application thereof, molecular genetic markers closely related to economic characters of meat ducks are screened and detected in the DNA level, and molecular bases are provided for rapid breeding of meat ducks.
Owner:江苏申中科技集团有限公司
A Method for Individual Breeding Value Assessment of Random Populations of Turbot
ActiveCN105755162BThe result is accurateAccurately estimate breeding valuesMicrobiological testing/measurementRandom populationPhysical Marking
The invention aims to provide a method for evaluating the individual breeding value of a turbot random population, so as to calculate an individual parentage coefficient under the condition of non-existence of physical pedigree, and further carry out genetic parameter evaluation. Pedigree recording is not needed in the method, but in traditional breeding, physical marking needs to be performed on a breeding population, and pedigree is recorded to evaluate an inter-individual genetic relationship, so that the workload is high. According to the method, the inter-individual genetic relationship can be directly evaluated by utilizing molecules, so as to eliminate the step of pedigree recording. Moreover, the inter-individual genetic relationship can be evaluated more accurately according to the method.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Screening method of goat COX II gene segment capable of serving as molecular marker and application thereof
InactiveCN102533746AExcellent genetic resourcesAccurately estimate breeding valuesMicrobiological testing/measurementDNA preparationBiotechnologyScreening method
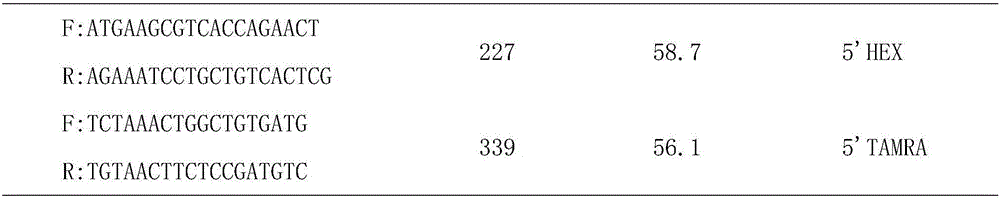
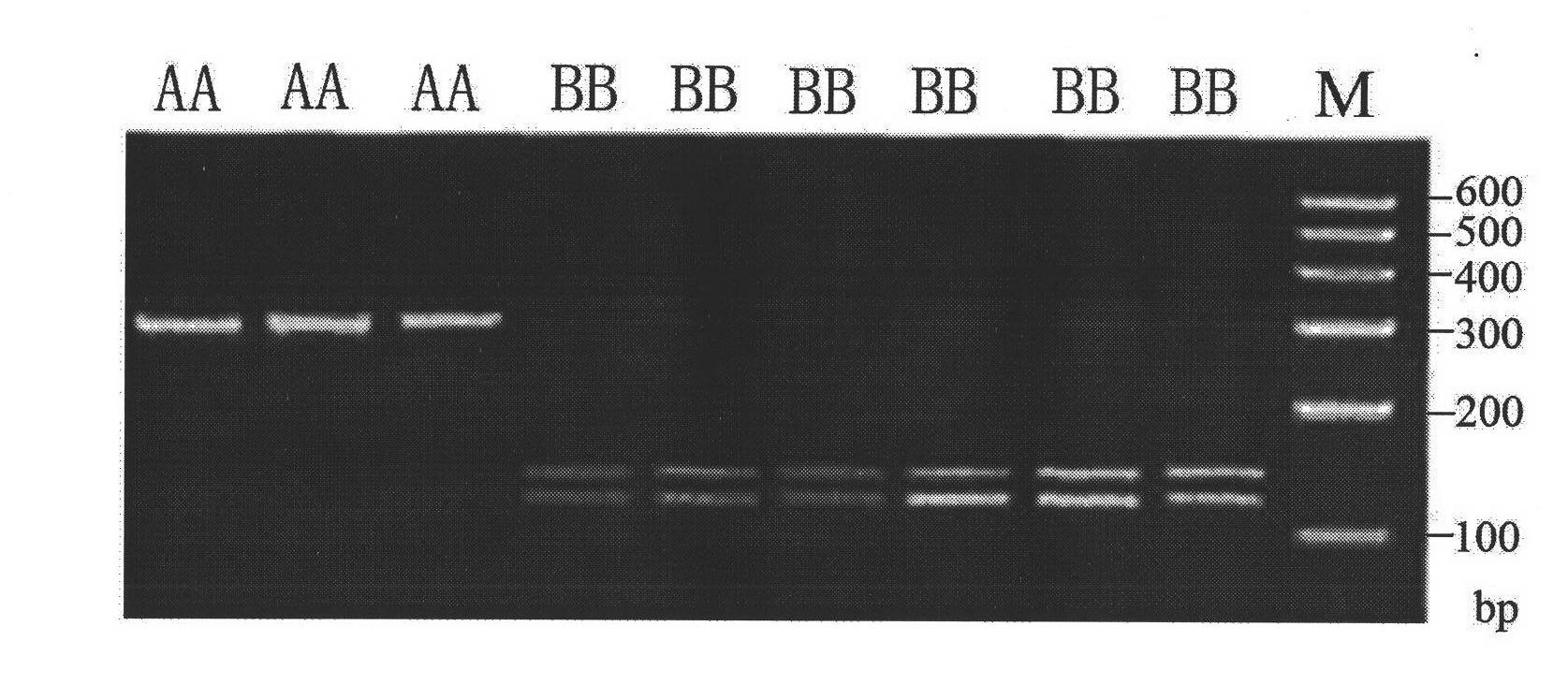
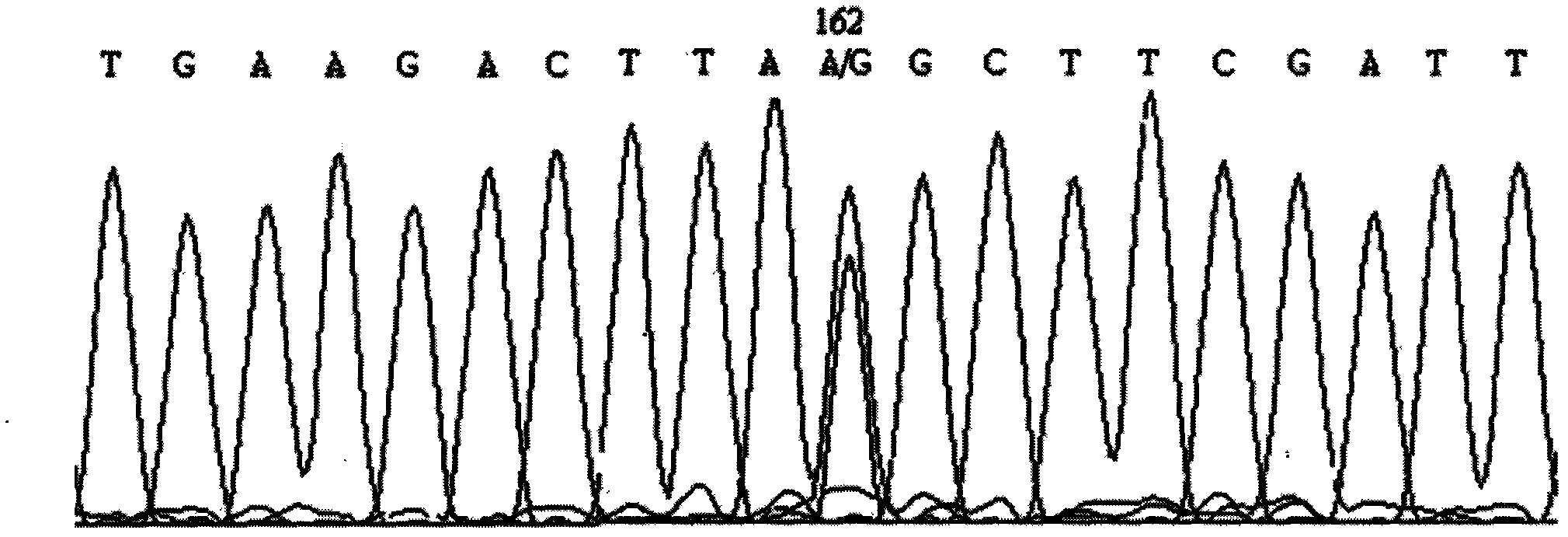
The invention discloses a screening method of a goat COX II gene segment capable of serving as a molecular marker and application thereof. The screening method comprises the following steps: by using constructed goat sperm pool DNA (deoxyribonucleic acid) as a template, using a designed specific primer P1 set a corresponding program in the presence of Tag DNA polymerase, buffer, Mg<2+> and dNTPs (deoxyribonucleotide triphosphates), and carrying out PCR (polymerase chain reaction) amplification on the COX II gene; determining the size of the target segment according to an agarose gel electrophoretogram; sequencing to obtain sequence and mutant site information, wherein the result indicates that the amplification product has one base mutant; after digesting HindI II the P1 amplification product with restriction endonuclease, detecting the digested amplification segment by agarose gel electrophoresis; and analyzing the genotype and gene frequency of the P1 amplification product as well as the correlation of the sperm activities between the Anhwei white goat and Bohr goat, wherein the result indicates that the SNP (single nucleotide polymorphism) site of the COX II gene detected by the primer P1 can serve as a molecular marker for selecting goat sperm activity property, thereby providing a molecular marker for auxiliary selection of a goat sperm quality property marker.
Owner:ANHUI AGRICULTURAL UNIVERSITY +1
A Molecular Marker Method for Selecting Growth Traits of Goats Using Nucleolin Gene
ActiveCN107287301BAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementBoer goat breedExon
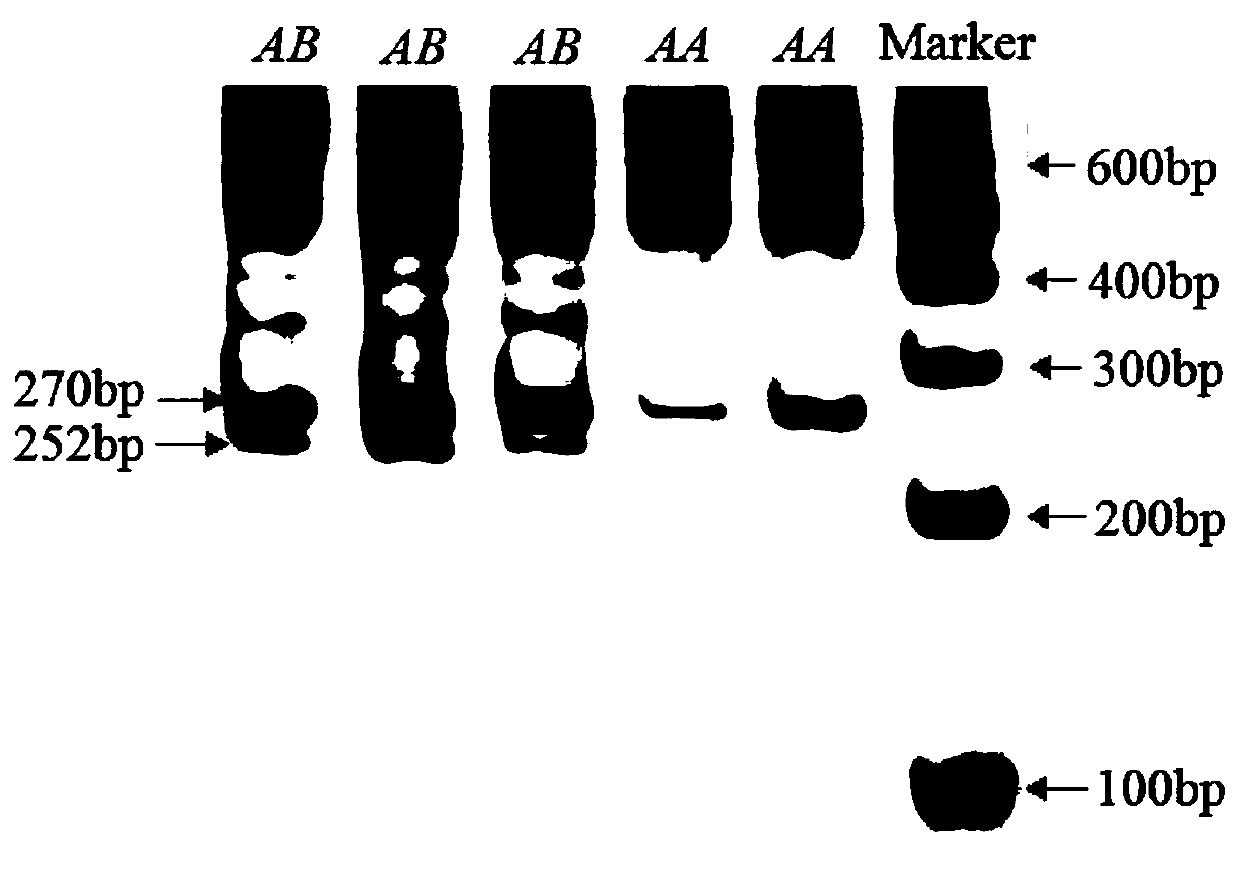
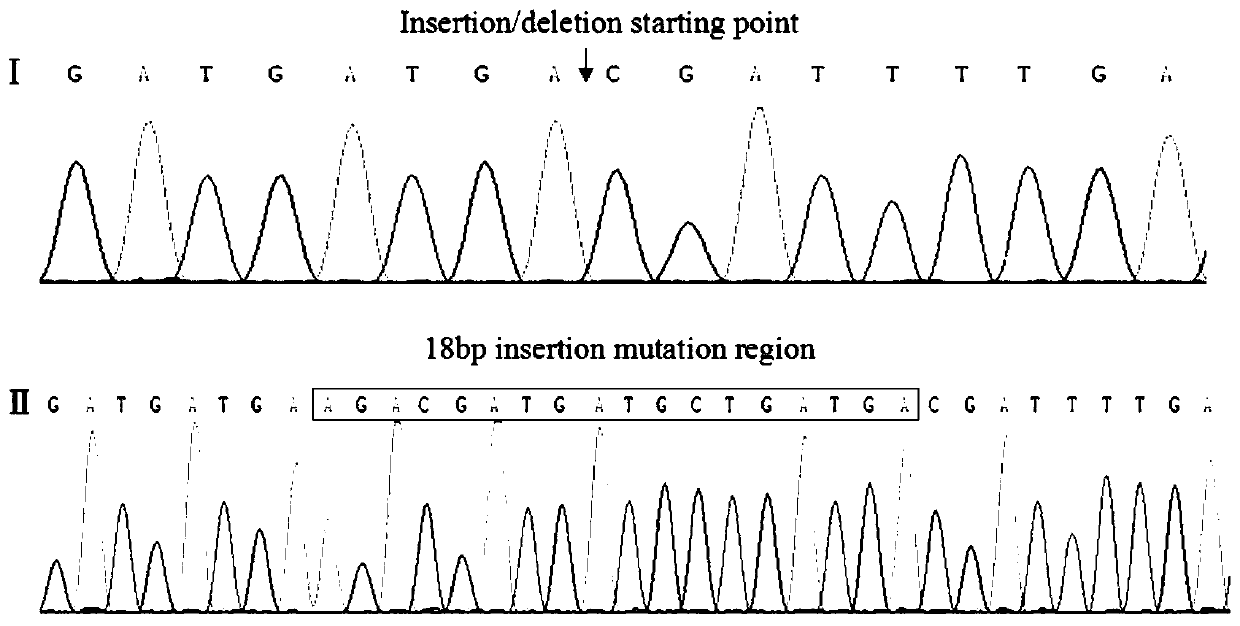
The invention discloses a molecular marking method selecting goat growth traits using nucleophosmin gene. According to the method, a goat gene group DNA sequence is used as a template, under the condition of existence of Taq DNA polymerase, buffer environment, Mg <2+>, and dNTPs, four pairs of primer are used for amplifying a Nucleophosmin (NPM) gene 5'UTR region and exon 1 under the condition of PCR condition, and the size of a target segment of an amplified product is determined according to agarose gel electrophoresis; an amplified site of primer P3 is found lacking of 18bp basic group through detection of PCR amplification products of primers P1-P4 by use of polyacrylamide gel electrophoresis, then genotyping and gene frequency analysis are performed on the amplification product of the primer P3, and associated analysis on the amplification production of the primer P3 and growth traits of a milk goat and a Boer goat in the central Shaanxi plain is performed. Results indicate that the SNPs site of the Nucleophosmin gene exon 1 detected by the primer P3 can be used as a molecular mark for goat growth trait selection.
Owner:NORTHWEST A & F UNIV
A method for breeding milk-producing traits of dairy goats using double-gene polymerization effect
InactiveCN103468819BAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementAnimal husbandryGene typeGenotype
The invention discloses a method for selecting milk production characters to breed a milk goat by utilizing the double gene polymerization effect. Milk goat genome DNA serves as a template, two pairs of primers are used for amplifying the exon 9 of a gene of a PRLR and the intron 1 of a gene of an ELF5 respectively, agarose gel electrophoresis with the concentration of 1.5% is used for judging the sizes of the amplified products, the locus mutations of the amplified products of the two pairs of primers are screened based on the DNA sequencing technique, then, the agarose gel electrophoresis with the concentration of 3.5% is used for carrying out genetic typing and gene frequency analyzing on SNPs of the two locuses of the gene of the PRLR and the gene of the ELF5, the relation between different gene type combinations and the milk production characters is analyzed, and the optimal gene combination is screened out.
Owner:NORTHWEST A & F UNIV
The method and application of detection of insertion-deletion polymorphism of sheep fth-1 gene by pcr-sscp
ActiveCN106701930BExcellent genetic resourcesAccurately estimate breeding valuesMicrobiological testing/measurementSingle-strand conformation polymorphismElectrophoreses
The invention discloses a method for detecting insertion-deletions polymorphism of sheep FTH-1 (ferritin heavy polypeptide1) genes by utilizing PCR-SSCP (polymerase chain reaction-single strand conformation polymorphism) and application thereof. The method comprises the following steps: taking to-be-detected sheep whole genome DNA containing FTH-1 genes as a template, performing PCR amplification on the sheep FTH-1 gene segments, and performing detection and genotyping on the PCR product by utilizing the SSCP technology; and identifying the 639th locus insertion-deletions polymorphism of the sheep FTH-1 genes according to electrophoretic results. The method is a method for screening and detecting a molecular genetic marker which is closely related to reproductive performances of sheep on the DNA level so as to be used for assistant selection and molecular breeding of the sheep and acceleration of sheep stock breeding.
Owner:甘肃润牧生物工程有限责任公司
Molecular marking method of using neuroendocrine factor genes to select kidding characters
InactiveCN101906480BAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementBase JGene selection
The invention discloses a molecular marking method of using neuroendocrine factor genes to select kidding characters. The method comprises the following steps of: using a goat genomic DNA sequence as a template; amplifying Kisspeptin gene intron 2 by using a primer P1 under the PCR condition in the presence of TaqDNA polymerase, buffering environment, Mg2+, dNTPs, and then judging the size of thetarget fragment by agarose gel electrophoresis; detecting the PCR application product of the primer P1 by polyacrylamide gel electrophoresis, finding that the amplification SNPs of the primer P1 has one-base mutation and 8bp-base deletion, and then carrying out genotyping and gene frequency analysis on the amplification product of the primer P1, and the correlation analysis with the kidding characters of Guanzhong diary goat and Xinong Saanen dairy goat. Shown by experiments, the SNPs of the Kisspeptin gene intron 2 detected by the primer P1can be used as molecular marker of the selection of kidding characters.
Owner:NORTHWEST A & F UNIV
The method and application of detection of single nucleotide polymorphism of sheep pcnp gene by pcr-rflp
ActiveCN106755371BAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementNucleotideGel electrophoresis
The invention discloses a method for detecting single nucleotide polymorphism of a sheep PCNP (PEST-Containing Nuclear Protein) gene by using PCR-RFLP (Polymerase Chain Reaction-Restriction Fragment Length Polymorphism) and application of the method. The method comprises the following steps: carrying out PCR amplification on a sheep PCNP gene segment by taking a sheep whole genome DNA to be measured as a template and taking a primer pair P as a primer; after a PCR amplification product is digested by a restriction enzyme SPhI, carrying out agarose gel electrophoresis on the digested amplified segment; authenticating the single nucleotide polymorphism of the 5019th site of the sheep PCNP gene according to an electrophoresis result. As a base mutation site is closely associated with the kidding number of sheep reproduction traits, the method is a method for detecting molecular genetic markers closely associated with the sheep reproduction traits at the DNA level, and can be used for auxiliary selection and molecular breeding of sheep and increasing the improved variety breeding speed of the sheep.
Owner:甘肃润牧生物工程有限责任公司
Method for selecting molecular marker for goat yeaning traits
InactiveCN101899526BAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementGel electrophoresisBoer goat
The invention discloses a method for selecting a molecular marker for goat yeaning traits, which comprises the following steps of: taking a goat genome DNA sequence as a template, amplifying KITL gene introns 1 and 6 by using primers P1 and P2 under a PCR condition in the presence of TaqDNA polymerase, buffer environment, Mg2+ and dNTPs respectively, and judging the size of a destination fragmentaccording to an agarose gel electrophoresis result; digesting the PCR amplification product of the primer P1 by using restriction enzyme CviAII, and then detecting the enzyme digested amplification fragment by using polyacrylamide gel electrophoresis, wherein the amplification product of the primer P1 has mutation of two basic groups; detecting the PCR amplification product of the primer P2 by adopting the polyacrylamide gel electrophoresis, wherein the amplification product of the primer P2 has mutation of one basic group; and then performing gene analysis and gene frequency analysis on the amplification products of the primers P1 and P2, and performing association analysis between the amplification products and the yeaning numbers of Guanzhong dairy goats, western Saanen dairy goats andBoer goats, wherein the analysis results show that an SNPs site of the KITL gene, detected by the primers P1 and P2, can be taken as the molecular marker for goat yeaning trait selection.
Owner:NORTHWEST A & F UNIV
A method for detecting single nucleotide polymorphism of sheep kitlg gene and its application
ActiveCN106498078BAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementKITLG GeneLitter
The invention discloses a method for detecting single nucleotide polymorphism of a sheep KITLG gene and application of the method. The method comprises the following steps: carrying out PCR amplification on fragments of the sheep KITLG gene in a manner of taking a whole-genome DNA of sheep to be detected as a template and taking a primer pair P as primers; digesting a PCR amplification product with restriction endonuclease SspI, and then, carrying out agarose gel electrophoresis on the amplified fragments subjected to digestion; identifying the polymorphism of a 47569-th base of the sheep KITLG gene according to electrophoresis results. The mutant site of the base is closely associated with reproduction traits, i.e., litter size of the sheep, so that the method is a method for detecting molecular genetic markers, which are closely related to the reproduction traits of the sheep, from a DNA level, and can be applied to the assisted selection and molecular breeding of the sheep, and thus, the good-variety breeding speed of the sheep is accelerated.
Owner:天津奥群羊业研究院有限公司
Single nucleotide polymorphism of ox TAS1R2 gene and detecting method thereof
InactiveCN101962683AAccurate estimateImprove selection efficiencyMicrobiological testing/measurementMolecular breedingPolymerase chain reaction
The invention discloses a single nucleotide polymorphism of an ox TAS1R2 gene and a detecting method thereof. The detecting method of the single nucleotide polymorphism of the gene comprises the following steps of: with an ox total genome DNA to be detected, which comprises a TAS1R2 gene, as a template and with a primer pair P as a primer, carrying out PCR (Polymerase Chain Reaction) amplification on the ox TAS1R2 gene; after denaturing the product, carrying out polyacrylamide gel electrophoresis on a denatured amplification fragment; and identifying the 288bp base polymorphism of the ox TAS1R2 gene according to an electrophoresis result. The method is used for screening and detecting molecular genetic markers which are closely relevant to ox meat producing characters at a DNA level to beused for the assisted selection and molecular breeding of oxen and improving the breeding speed of fine breeds of the oxen.
Owner:XUZHOU NORMAL UNIVERSITY
Detection method and application of molecular markers for single nucleotide polymorphism of foxo1 gene in Qinchuan cattle
InactiveCN103233001BAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationNucleotideElectrophoresis
Owner:NORTHWEST A & F UNIV
A Molecular Marker Method for Selecting Goat Litter Size Using a Delinked Metalloprotease Gene Containing a Thrombospondin Motif
ActiveCN106566881BAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementMetalloprotease GeneMetalloproteinase Gene
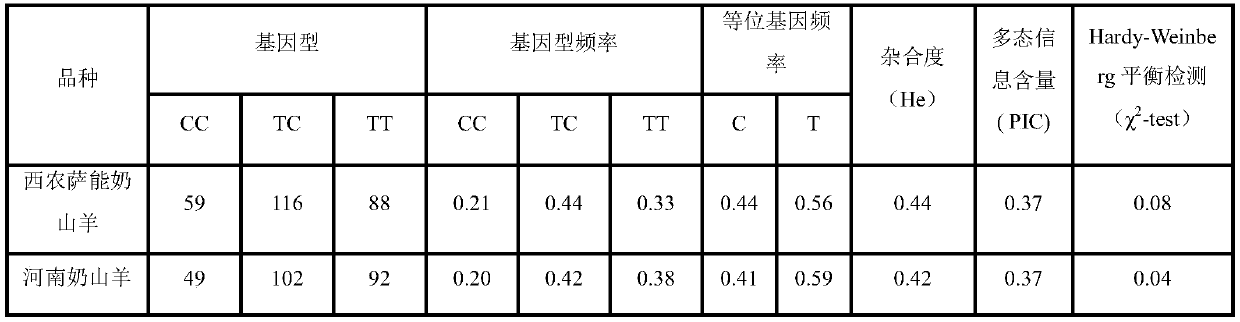
The invention discloses a molecular marking method for selecting the goat kidding number by using a de-linked metalloprotease gene containing a thrombin-sensitive protein die body. The method adopts the DNA sequence of a goat genome as a template. With the presence of a taq DNA polymerase, a buffer environment, Mg2+ and dNTPs, the gene exon 2 of a de-linked metalloprotease (ADAMTS 1) containing the thrombin-sensitive protein die body is amplified with the presence of a primer P1 in the PCR condition. Based on the DNA sequencing technique, the single nucleotide polymorphism (SNP) of the exon 2 is detected. Gene sequencing results show that, the amplification sites of the primer P1 contains one base mutation, and then the amplified product of the primer P1 is subjected to genetic typing and gene frequency analysis. Meanwhile, the association analysis is conducted on the kidding traits of Henan dairy goats and the kidding traits of Xinong Saanen dairy goats. Results show that, the SNP sites of the gene exon 2 of the de-linked metalloprotease (ADAMTS 1), detected by the primer P1, can be used as molecular markers for selecting the kidding number traits of goats.
Owner:河南省动物卫生监督所
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com