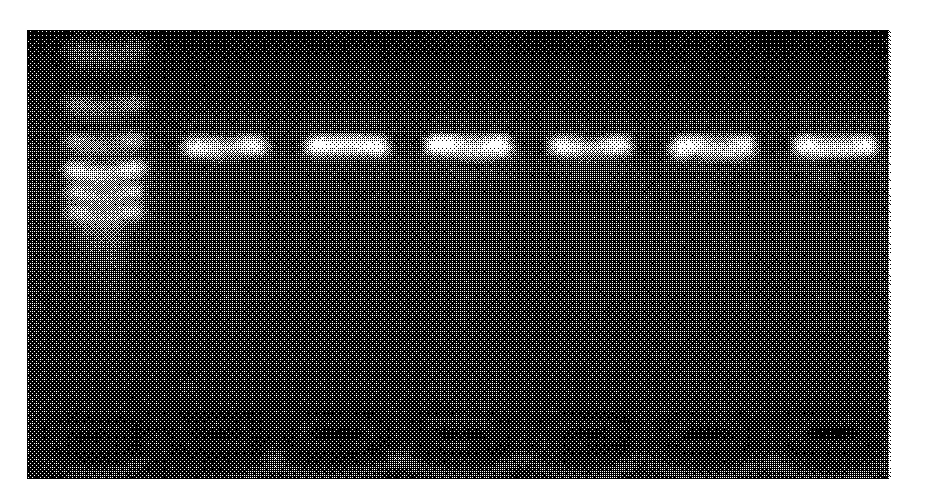Method for detecting base mutation polymorphism of goat gonadotropin releasing hormone and growth differentiation factor 9
A growth differentiation factor and gonadotropin technology, applied in the field of molecular genetics, can solve the problems of expensive detection, cumbersome operation, long experimental process, etc., and achieve the effect of speeding up the breeding progress and improving the selection efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
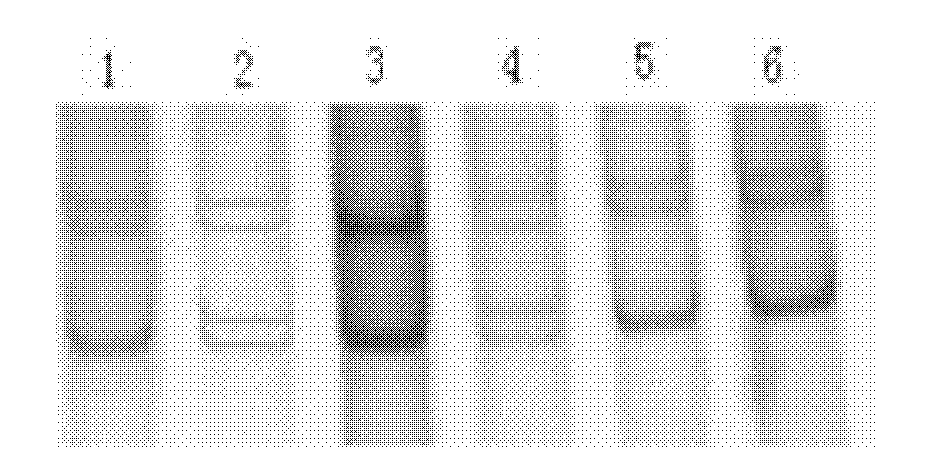
[0015] , GnRH gene exon 4 (including part of intron 3 and 3'untranslated region) and GDF9 gene exon 2 PCR amplification and polymorphism detection 1. Collection and processing of goat blood samples Take goat blood samples Add 0.2 μL of ACD (citric acid 2.4g; trisodium citrate 6.6g; glucose 7.35g; dilute to 50mL, autoclave) to anticoagulate, slowly invert 3 times, put in ice box, and store at -80°C spare.
[0016] In this example, a total of 720 ewe blood samples from 2 goat populations were used, specifically: 406 blood samples of Xinong Saanen sheep, collected from Qianyang Saaneng sheep breeding farm in Shaanxi Province; 314 blood samples of Boer goats, collected from Shaanxi Province Boer Goat Breeding Farm in Linyou County and Luonan Branch of Yangling Jinkun Company in Shaanxi Province.
[0017] 2. Extraction and purification of genomic DNA from blood sample 1) Thaw the frozen blood sample at room temperature, transfer 500 μL to a 1.5 mL Eppendorf tube, add an equal volu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com