Molecular marking method for indicating Qinchuan cattle meat tenderness and water-holding capacity by utilizing THRSP (thyroid hormone responsive spot) genes
A technology of molecular markers and bovine muscle, applied in biochemical equipment and methods, microbiological measurement/inspection, etc., can solve the problems of poor muscle fat deposition ability, poor breast development of cows, low milk production, etc., and achieve broad development The effect of prospect and profit margin, high precision and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] This example is used to illustrate the acquisition of whole blood genomic DNA of Qinchuan cattle.
[0027] (1) Thaw the frozen blood sample at room temperature, add about 3mL whole blood into a 7mL centrifuge tube, add an equal volume of PBS buffer, mix for 10min, centrifuge at 12000rpm for 5min, discard the supernatant, and repeat twice;
[0028] (2) Add 2mL STE, mix for 5-10min, and bathe in 37℃ water for 1h;
[0029] (3) Add proteinase K (20mg / mL) to a final concentration of 100μg / μL, and mix well;
[0030] (4) Add 70 μL of SDS, shake gently in a 56°C water bath, and digest overnight until viscous blood clots cannot be seen;
[0031] (5) Add an equal volume of Tris saturated phenol, cap the tube tightly, slowly mix it back and forth for at least 10 minutes, and centrifuge at 12000 rpm and 4°C for 10 minutes;
[0032] (6) Transfer the supernatant to a new sterilized centrifuge tube, and repeat the extraction once with Tris saturated phenol;
[0033] (7) Then add an...
Embodiment 2
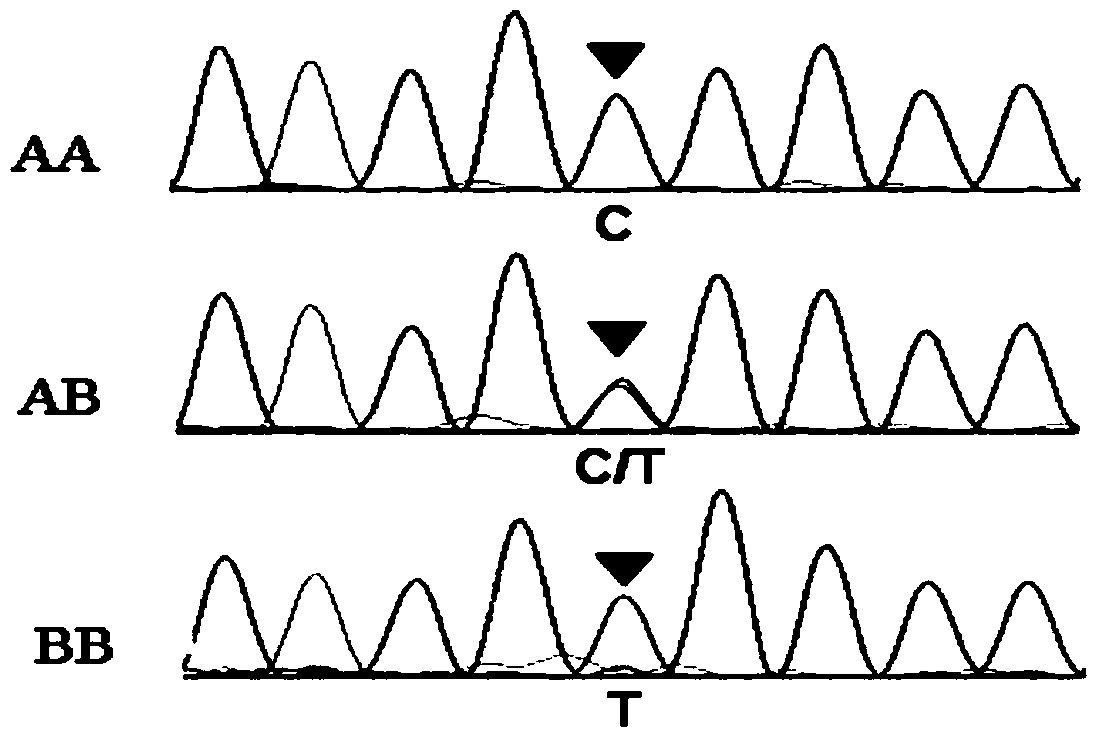
[0040] This example is used to illustrate PCR-SSCP analysis.
[0041] Referring to the sequence of bovine THRSP gene provided in GeneBank (accession number: No. AY656814), primers were designed using Primer5.0 software. The primer sequences are as follows:
[0042] THRSP-F: 5'-3': CGCCTCCGATCTCTACAACT (SEQ ID No. 1)
[0043] THRSP-R: 5'-3': CCAGCTTCAGTCTCAGCTCC (SEQ ID No. 2)
[0044] The Qinchuan cattle whole blood genomic DNA obtained in Example 1 was used for PCR reaction using the above primers. The reaction conditions were 95°C, pre-denaturation for 5 min; 95°C, denaturation for 30 sec, 56.5°C, renaturation for 30 sec, 72°C extension for 30 sec, a total of 35 cycles; 72°C extension for 10 min.
[0045] After the reaction, the PCR reaction solution (5 μL) was taken for agarose gel electrophoresis to detect the PCR product.
[0046]Take 5 μL of the PCR product and mix it with 8 μL of loading buffer (deionized formamide with a volume fraction of 98%, bromophenol blue wit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com