Method for improving yield of arginine by mutation of Corynebacterium crenatum N-acetyl glutamic acid kinase
A technology of glutamic acid kinase and acetylglutamate, which is applied in the field of arginine production key enzyme - N-acetylglutamate kinase, to produce arginine, and genetically engineered recombinant corynebacterium bacillus to produce arginine, Ability to solve problems such as genetic background being in the blank state
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
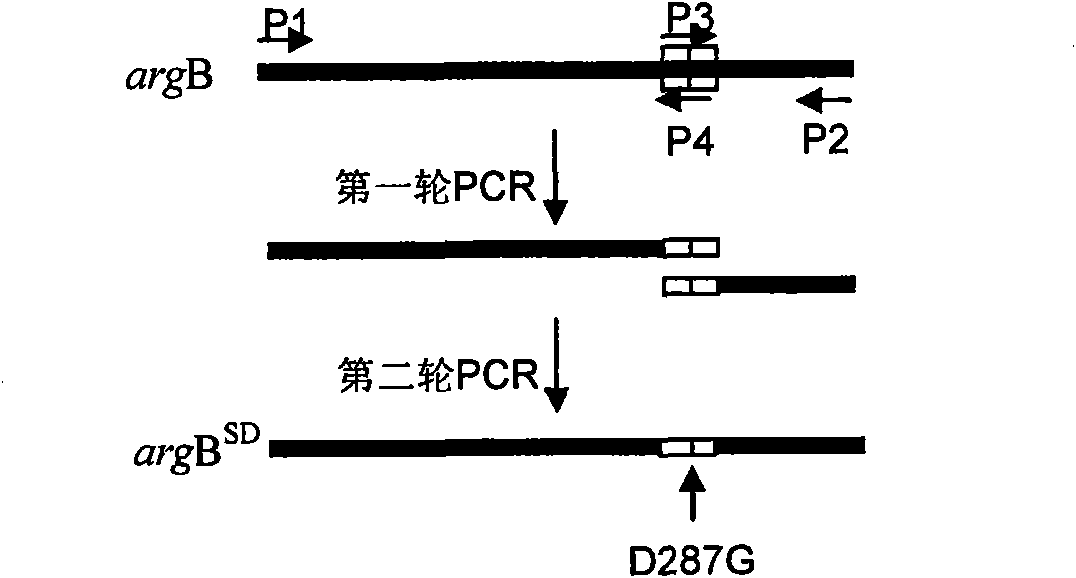
[0022] Example 1. Cloning of mutant argB SD Gene
[0023] The wild-type argB gene was amplified by PCR program, and cloned into vector pET28a to generate plasmid pET28a-argB. The genomic DNA of Corynebacterium blunt tooth was used as a template, the oligonucleotides P1 and P2 described in SEQ ID NO: 3 and 4 were used as primers, and the PCR amplification parameters were denaturation at 94°C for 90s, annealing at 55°C for 1min, and 72°C Extend 1.5min, 35 cycles. The resulting gel recovered product was ligated with pMD18-T vector and sequenced. The sequencing result is the nucleotide sequence shown in SEQ ID NO:2. The mutated argB gene was obtained through two rounds of PCR by overlapping PCR. The parameters of the two rounds of PCR amplification were denaturation at 94C for 30s, annealing at 52C for 30s, extension at 72C for 30s, and 25 cycles. The plasmid pMD18-T-argB was extracted as the template for the first round of site-directed mutagenesis, and PCR amplification was...
Embodiment 2
[0024] Example 2. Build with argB SD Recombinant Escherichia coli mutants with mutant genes
[0025] The argB obtained through two rounds of PCR in embodiment 1 SD The mutated gene was digested with EcoRI and SalI, and argB was recovered from the gel SD Fragment, connect it with the plasmid pET-28a cut by the same restriction enzyme, transform in Escherichia coli BL21 (DE3), screen the positive transformant, and obtain the positive transformant BL21 / pET-28a-argB SD . Select positive transformants and inoculate them in LB liquid medium containing 50 μg / mL kanamycin, culture overnight at 37°C on a shaker, and transfer to a 250mL Erlenmeyer flask containing 50mL medium the next day at a 1% inoculum size Continue to culture for 4h to the logarithmic growth phase, add IPTG (final concentration: 1mmol / L), and induce expression overnight at 16°C. Using the 6·HisTag coding sequence contained in the Escherichia coli expression vector pET-28a, the acetylglutamate kinase NAGK can be ...
Embodiment 3
[0025] The argB obtained through two rounds of PCR in embodiment 1 SD The mutated gene was digested with EcoRI and SalI, and argB was recovered from the gel SD Fragment, connect it with the plasmid pET-28a cut by the same restriction enzyme, transform in Escherichia coli BL21 (DE3), screen the positive transformant, and obtain the positive transformant BL21 / pET-28a-argB SD . Select positive transformants and inoculate them in LB liquid medium containing 50 μg / mL kanamycin, culture overnight at 37°C on a shaker, and transfer to a 250mL Erlenmeyer flask containing 50mL medium the next day at a 1% inoculum size Continue to culture for 4h to the logarithmic growth phase, add IPTG (final concentration: 1mmol / L), and induce expression overnight at 16°C. Using the 6·HisTag coding sequence contained in the Escherichia coli expression vector pET-28a, the acetylglutamate kinase NAGK can be purified by Ni-NTA column purification method. The protein activity of the purified NAGK was de...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com