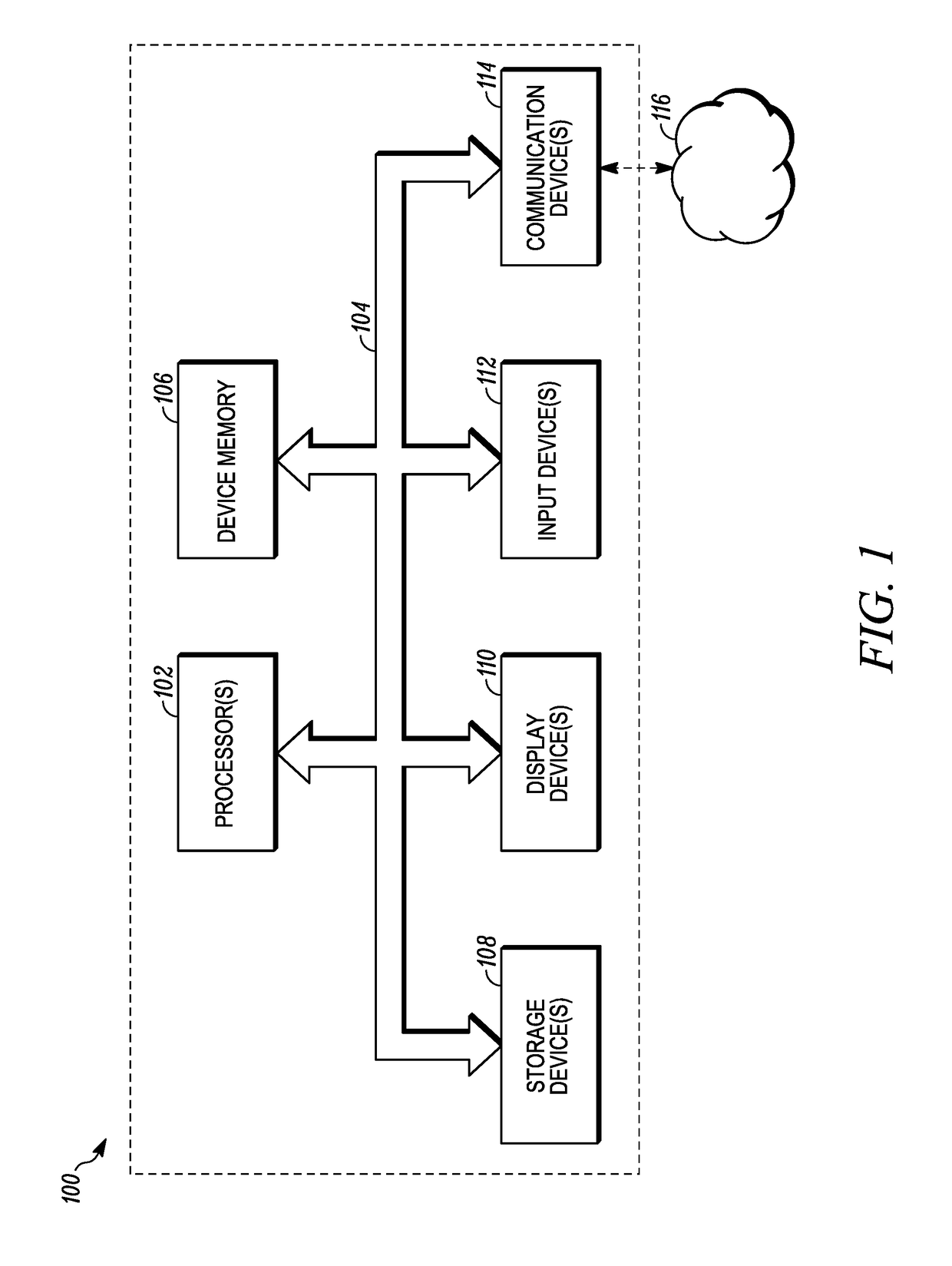
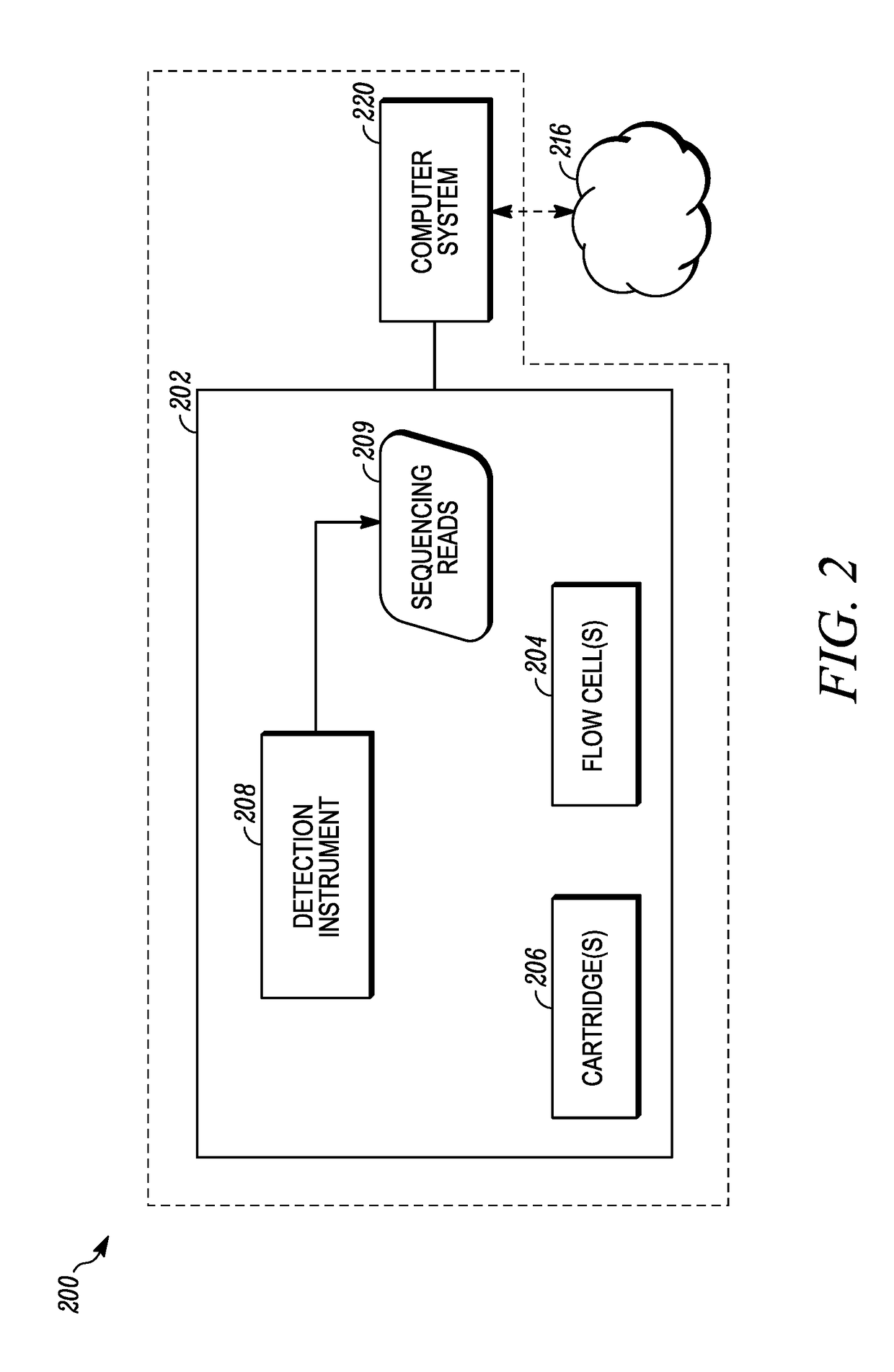
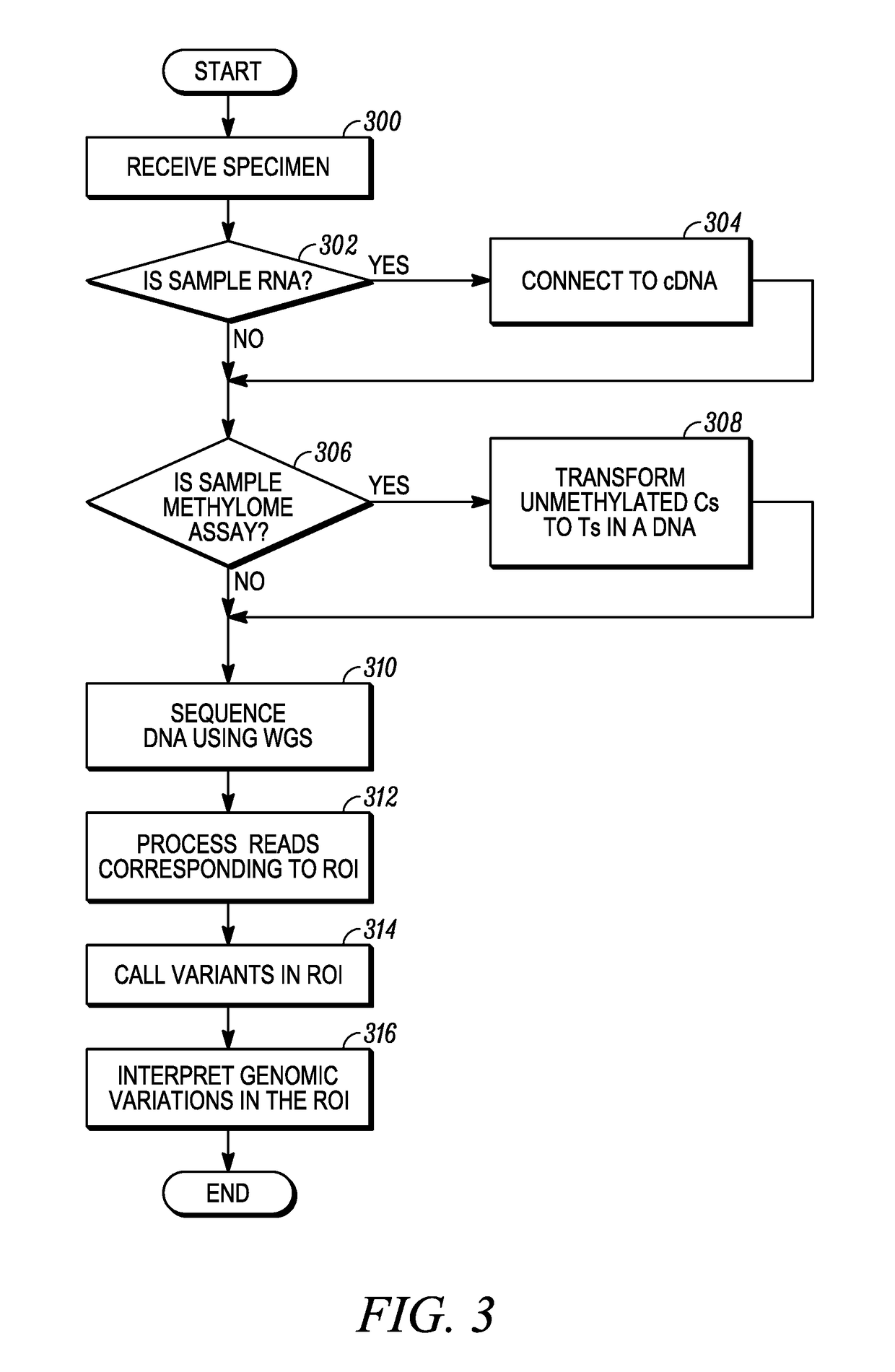
Patents
Literature
61 results about "Dna identification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
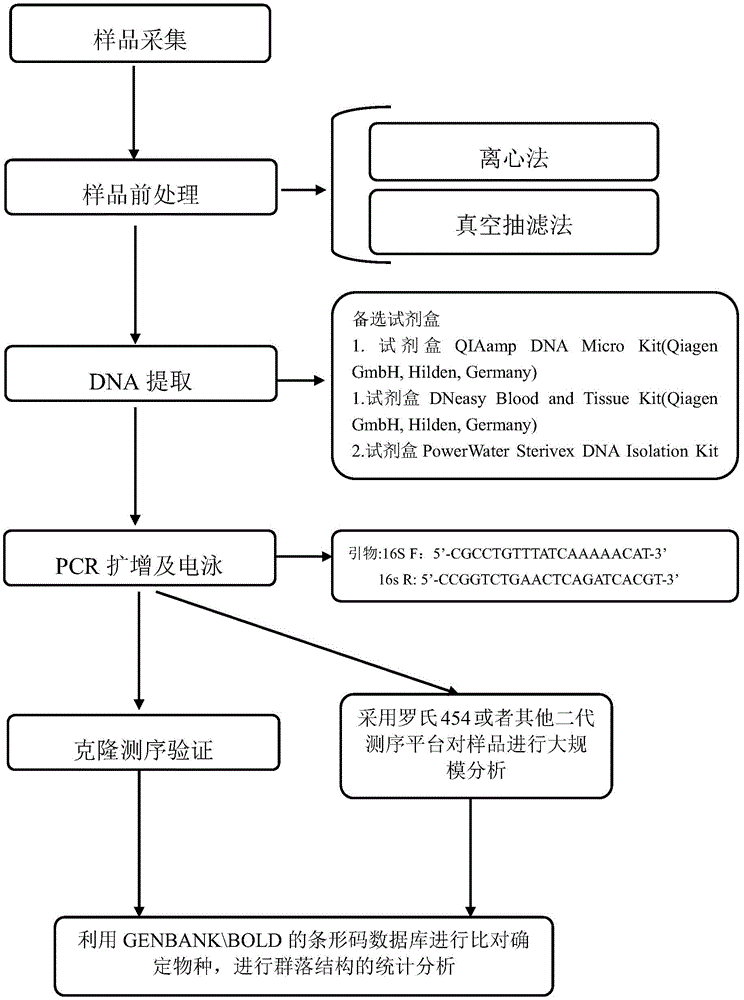
Environment DNA identification method for fish community structure researching
ActiveCN104531879AProtected speciesSave human and financial resourcesMicrobiological testing/measurementCommunity structureNucleotide sequencing
The invention relates to an environment DNA identification method for fish community structure researching. The method is characterized by comprising the steps that 1, water samples are collected according to the size of the water area where a fish community is located; 2, the water samples are processed, and total environment DNA of the processed water samples is extracted; 3, a 16s r DNA sequence with nucleotide sequences being SEQ ID NO:1 and SEQ ID NO:2 is adopted as a universal primer to carry out PCR amplification on the total environment DNA, and a PCR product is obtained; 4, gel electrophoresis is carried out on the PCR product, and gel with DNA is obtained; 5, gel cutting and clone sequencing are carried out on the gel with the DNA, and then blast comparison is carried out through GENBANK so as to determine whether the DNA is a fish sequence or not; 6, after it is determined that the DNA is the fish sequence, the next-generation sequencing technology is adopted for analyzing composition conditions of the PCR product on a large scale, and therefore the environment DNA fish composition and community structure are determined. The environment DNA identification method is quite easy, convenient and efficient and has the practical value.
Owner:SHANGHAI OCEAN UNIV
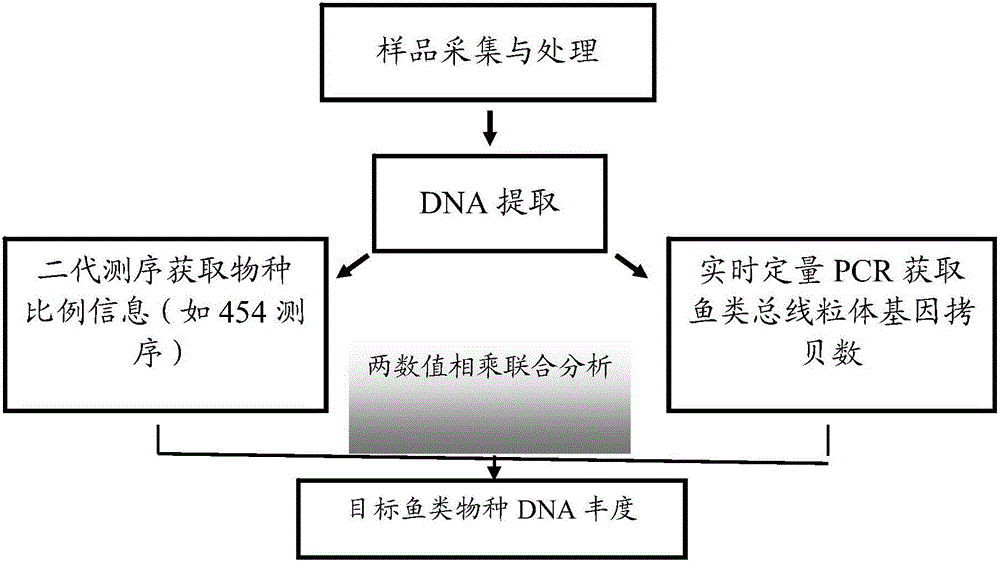
Conjoint analysis method for estimating DNA abundance of fishes based on environment DNA technology
ActiveCN105154564ASampling method is simpleProtect resourcesMicrobiological testing/measurementSequence analysisConjoint analysis
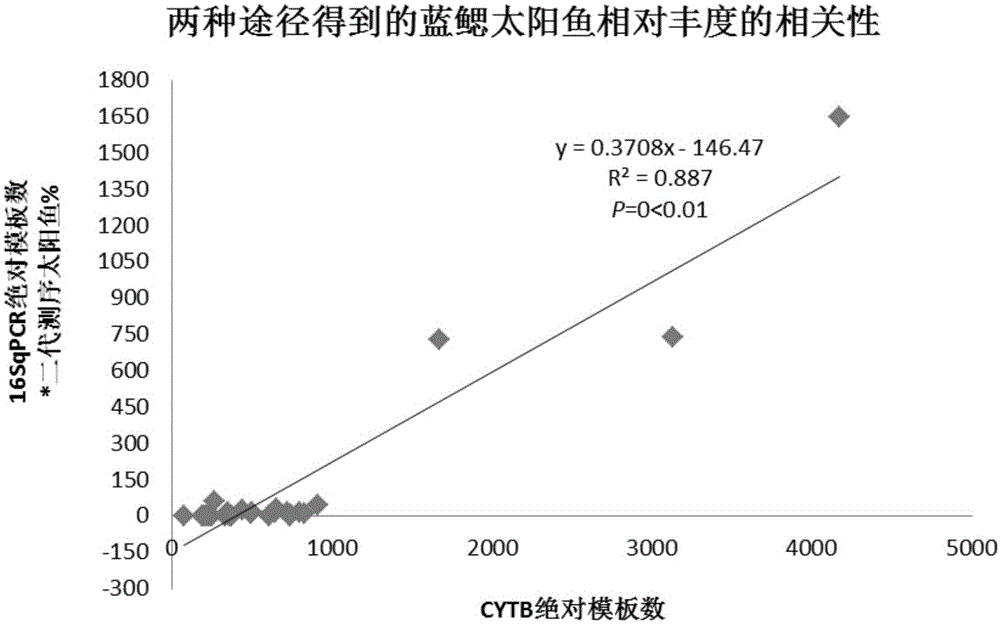
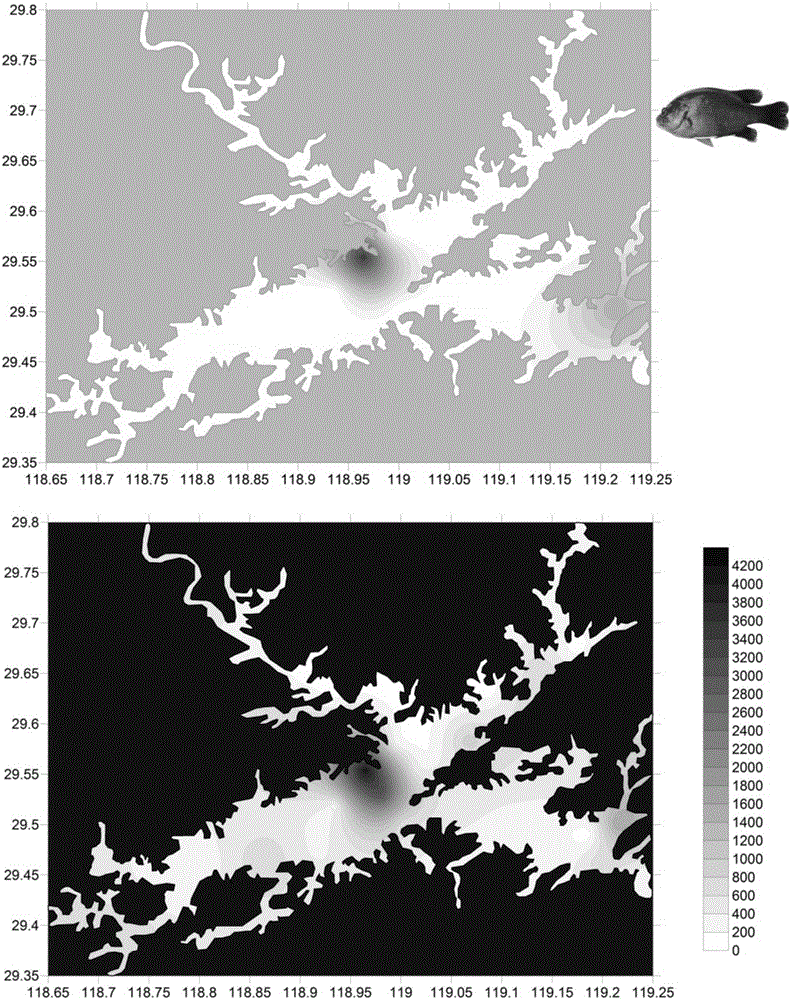
The invention relates to a conjoint analysis method for estimating the DNA abundance of fishes based on an environment DNA technology. The conjoint analysis method includes the steps that the total DNA abundance of the fishes is acquired through timed and quantified PCR; the species composition proportion of the fishes is acquired through 454 GS-FLX Titanium sequencing analysis; conjoint analysis of target species is performed through sequencing data and timed and quantified PCR data; a relative distribution diagram of mitochondrial DNA of the fishes in an environment is drawn through sufer 8.0 software. Based on the environment DNA identification technology, weather species exist and how many species exist can be analyzed just by collecting water samples without depending on fish species capturing, a sampling method is simple, and fish resources can be protected to the maximum degree; meanwhile, for some species which are rarely distributed and difficult to capture, the method is still more effective than traditional fishing investigation, the method can effectively solve the problem that effective species specificity primers are difficult to obtain when multiple closely related species exist in samples, and NDA abundance information of all species can be known just by conducting experiment once respectively through the two technologies.
Owner:SHANGHAI OCEAN UNIV
Method for breeding low temperature-resistant large yellow croakers
InactiveCN101606502AGood resistance to low temperature environmentClimate change adaptationPisciculture and aquariaAquatic productZoology
The invention belongs to the field of seawater aquaculture and particularly relates to a method for breeding and identifying low temperature-resistant large yellow croakers (also known as Pseudosciaena crocea). The invention mainly solves the status quo that a majority of large yellow croakers are not capable of accommodating to low-temperature culture currently; and the invention provides a method for breeding low temperature-resistant large yellow croakers by selecting large yellow croaker individuals capable of naturally resisting the low temperature and breeding on preferential basis, and provides a method for identifying low temperature-resistant large yellow croakers at the same time. The low temperature-resistant large yellow croakers are obtained through multi-generation breeding and culturing, observation-based identification and DNA identification.
Owner:ZHEJIANG OCEAN UNIV
Turtle shell DNA detection kit and identification method
InactiveCN103074433AMultiplex PCR identification method is simpleRapid multiplex PCR identification methodMicrobiological testing/measurementBioinformaticsDna identification
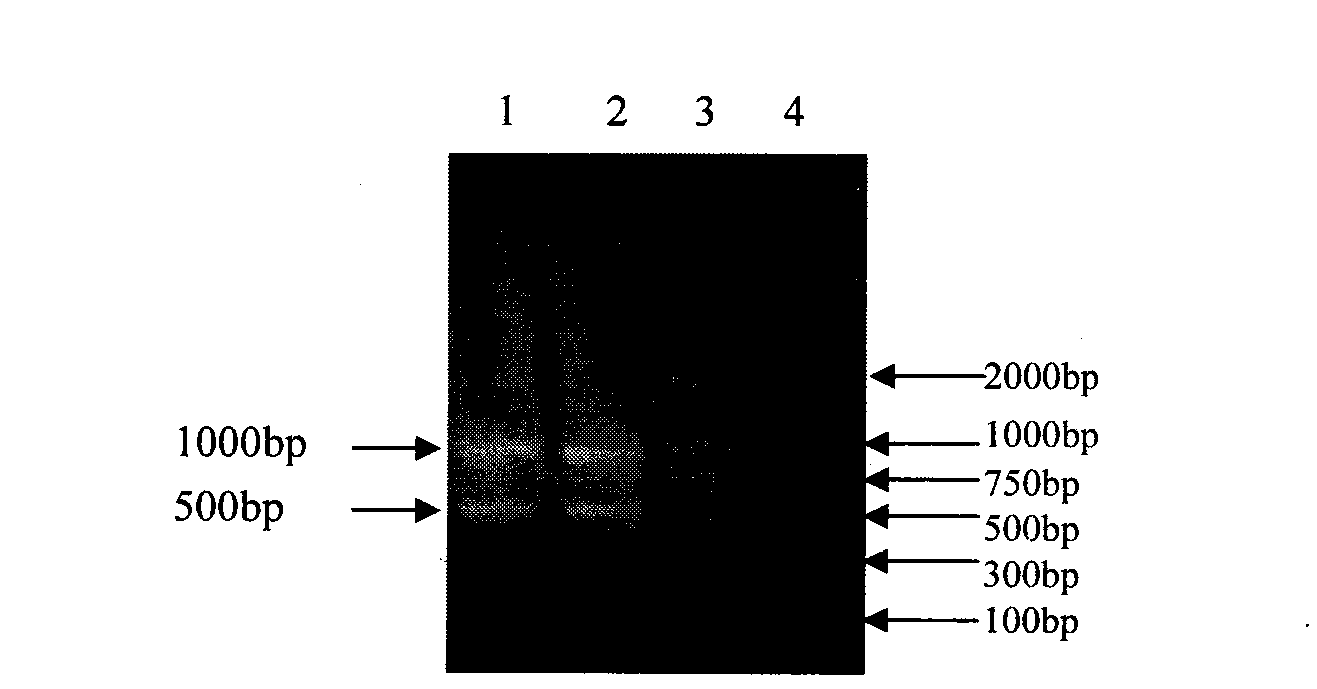
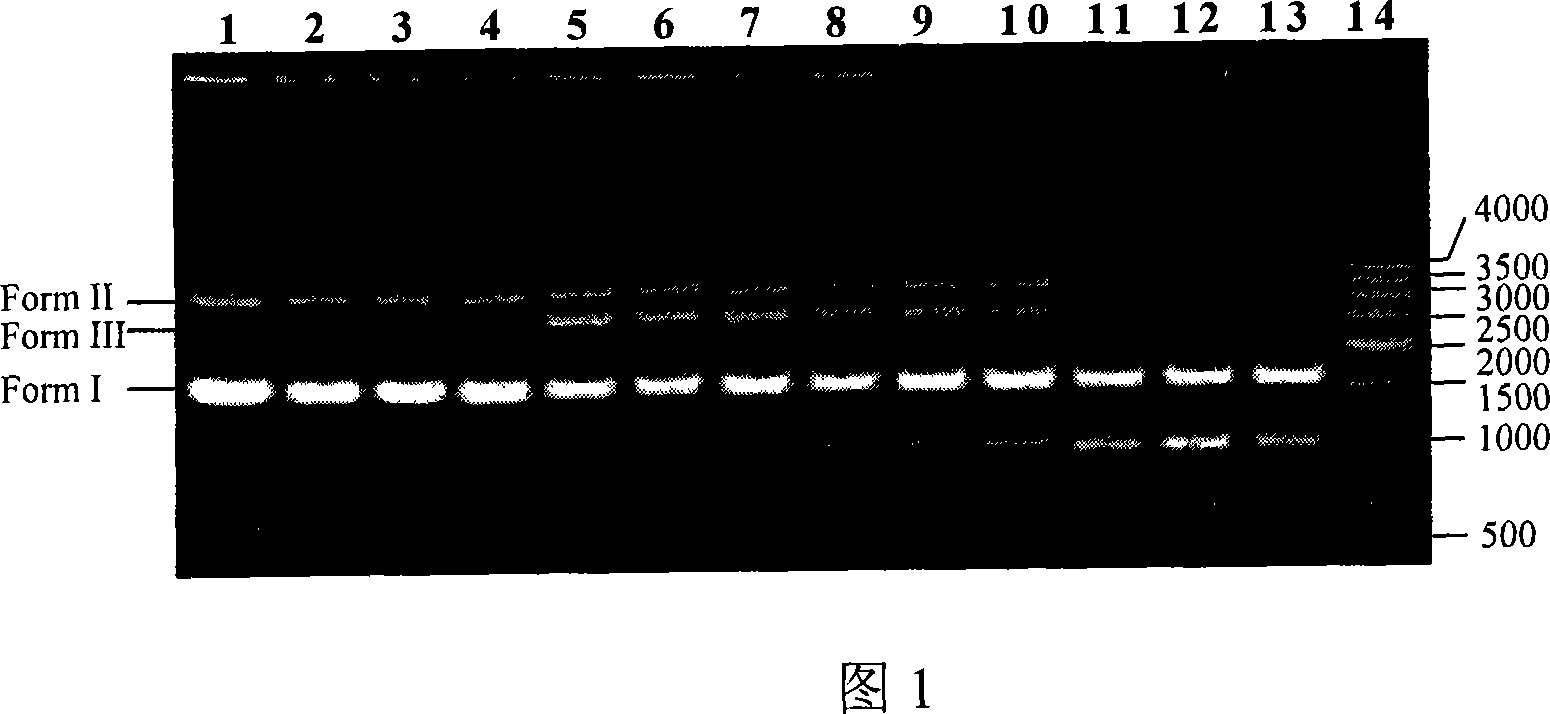
The invention relates to the technical field of traditional Chinese medicine identification, in particular to a turtle shell DNA (deoxyribonucleic acid) detection kit and an identification method. The turtle shell DNA detection kit comprises a sample pretreatment liquid, a decalcification liquid, a mitochondrial DNA extraction system, a PCR (polymerase chain reaction) system and a result observation system. The turtle shell DNA identification method comprises the steps of pretreating a detection sample, extracting mitochondrial DNA, conducting PCR primer design and synthesis, establishing PCR, and judging a result. A judgment standard is that a turtle shell is authentic if 1000bp and 500bp bands appear simultaneously, and the turtle shell is fake if only one or no band appears. The established multi-PCR identification method has the advantages of simplicity, convenience, quickness, reliable detection result and the like, and can differentiate and identify specificities of the turtle shell and the fake turtle shell accurately and simultaneously.
Owner:JILIN LEIBO TECH
Mercuric ion detection kit based on constant-temperature cascading nucleic acid amplification and detection method thereof
InactiveCN105256033ASimple designNo need to add system complexityMicrobiological testing/measurementMercuric ionFluorescence
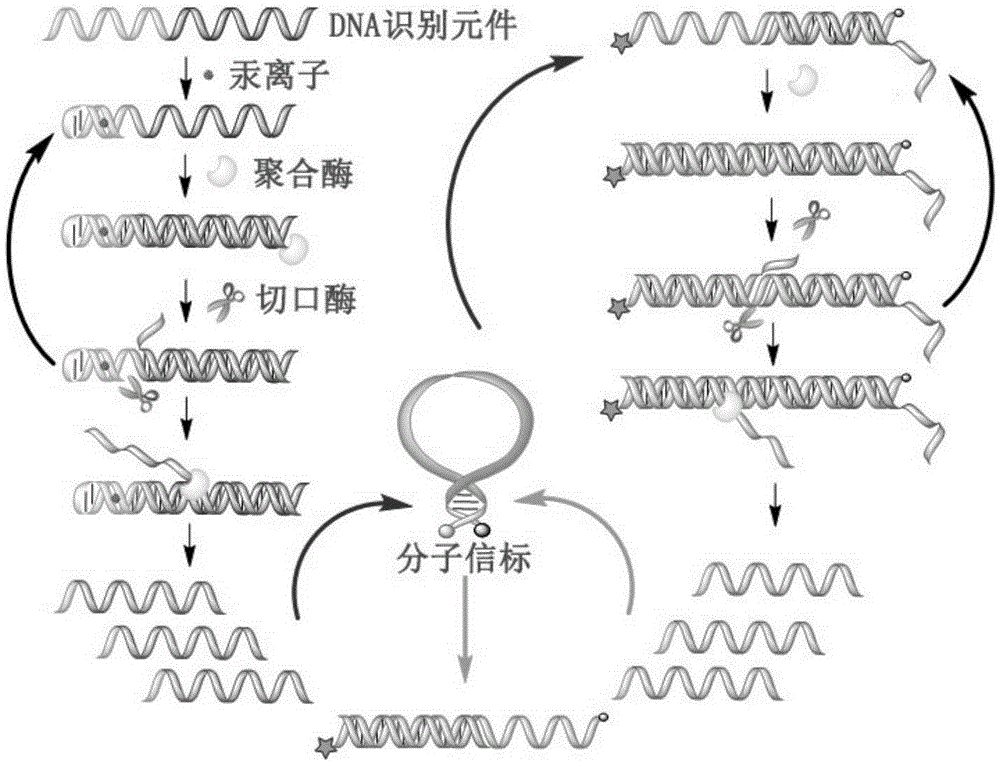
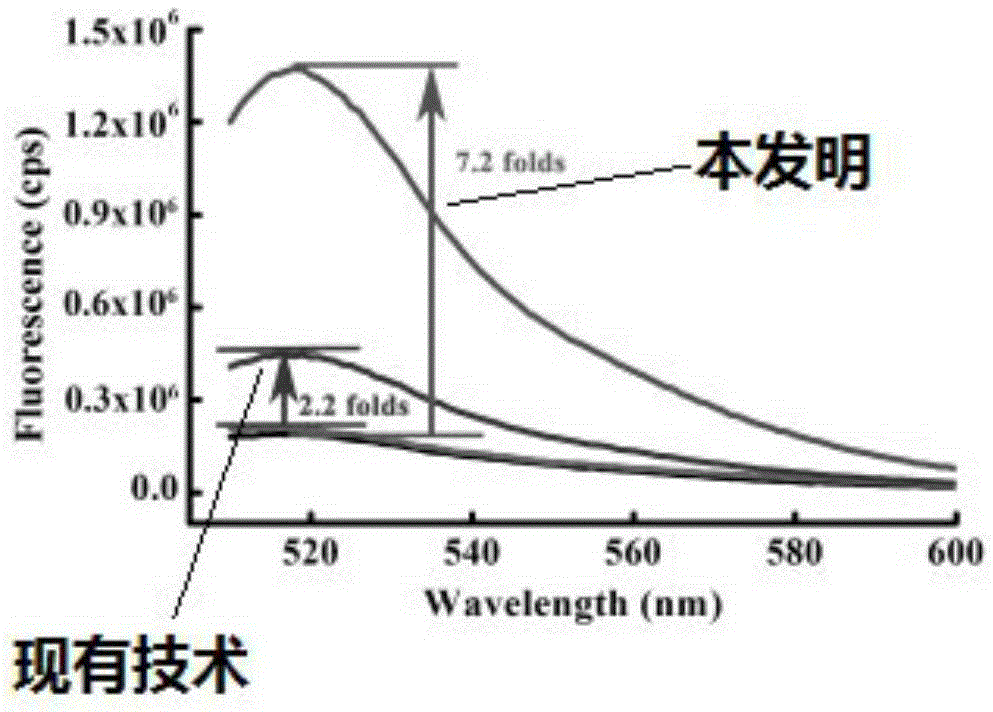
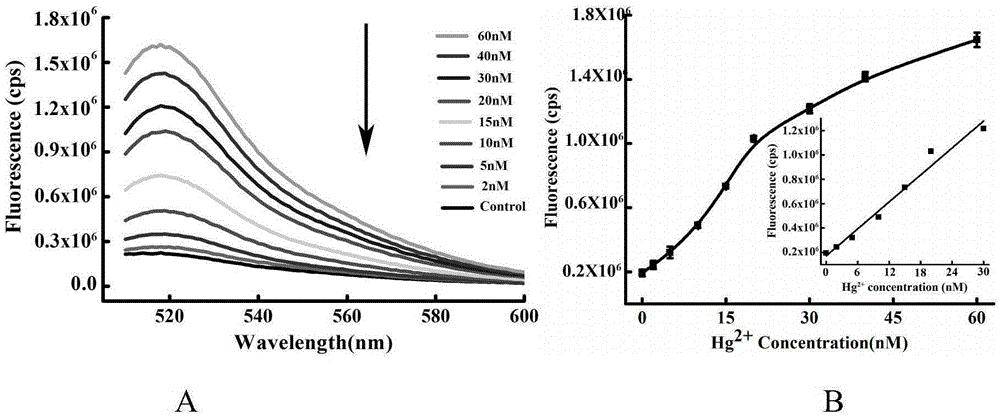
The invention provides a mercuric ion detection kit based on constant-temperature cascading nucleic acid amplification and a detection method thereof. Thymine-containing DNA identification elements are specifically bound with mercuric ions and then are folded to form an intra-molecular stem-loop structure; The 3' terminal of the DNA can be identified by polymerase, the DNA itself serves as a template to start strand displacement amplification reaction so as to produce a large amount of single-stranded DNA products; the single-stranded DNA products can open a restriction enzyme cutting site containing molecular beacon stem-loop structure to produce fluorescence signals; at the same time, the single-stranded DNA products can be also used as primers, and hybridization combined molecular beacons are used as templates to trigger secondary strand displacement amplification reaction, and released SDA products can form heteroduplexes with new molecular beacons so as to produce cascaded-amplified fluorescent signals. The detection method is high in sensitivity, ingeniously achieves cascading amplification of strand displacement amplification reaction without system complexity increase, is high in amplification efficiency and response speed and can achieve mercuric ion quantification within 30 minutes, and the detection limit is as low as 2 nM.
Owner:XI AN JIAOTONG UNIV +1
Mink heart DNA detection kit and identification method
InactiveCN101434990ASpecificity Accurate Simultaneous DistinctionThe identification method is simpleMicrobiological testing/measurementMinkPositive control
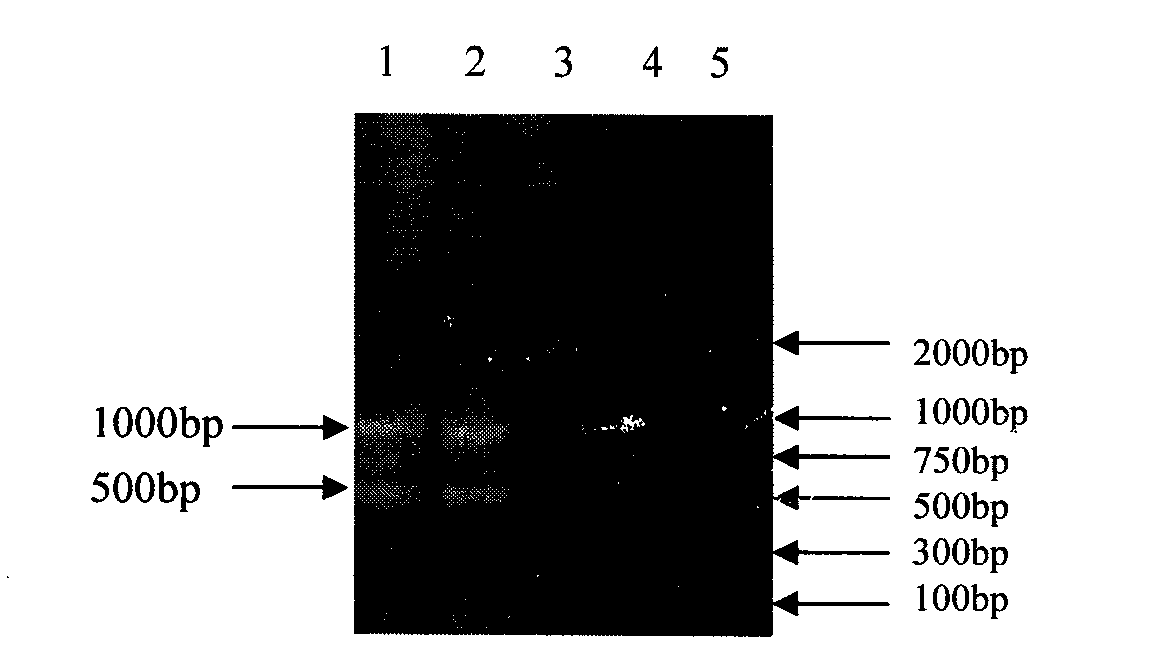
The invention relates to the Chinese medicine detection technology field and belongs to a mink heart DNA detection kit, comprising buffer solution, 12.5mM dNTP, one type of 0.1mM primer 1 or 2, Taq DNA polymerase, sample DNA to be detected, and the identification reaction system of double-distilled water; containing buffer solution, 12.5mM dNTP, one type of 0.1mM primer 1 or 2, Taq DNA polymerase, mink heart DNA, and the positive control reaction system of the double-distilled water; including buffer solution, 12.5mM dNTP, one type of 0.1mM primer 1 or 2, Taq DNA polymerase, mixture of chicken heart, duck heart, goose heart and rabbit heart DNA according to 1:1, and the negative control reaction system of the double-distilled water. The mink heart DNA identification method comprises steps such as designing mink heart mitochondrial DNA two-pair specific oligonucleotide primer, artificially synthetizing mink heart mitochondrial DNA two-pair specific oligonucleotide primer, fixing reaction procedures and result judgment, and the like; the method can simultaneously and accurately differ the specificity of various animal hearts which are easily mixed with the mink heat; and the identification method has the advantages of simplicity, rapidness, and reliable detection result, etc.
Owner:BEIHUA UNIV
Auxiliary optical device of attenuation total reflection surface enhanced infrared spectrometer for DNA analysis
InactiveCN102507444AAvoid interferenceKeep aliveColor/spectral properties measurementsUltrasound attenuationAnalysis dna
The invention relates to an attenuation total reflection surface enhanced infrared optical device for DNA identification and hybridization dynamics analysis. The attenuation total reflection surface enhanced infrared optical device comprises an attenuation total reflection surface enhanced infrared optical window in which an island-shaped nanometer gold film is plated on the surface of a silicon hemisphere, an optical table base, a fixed support region, an infrared light spectrum pool, a PDMS (Polydimethylsiloxane) sealing ring and the like, wherein the silicon hemisphere plated with the gold island-shaped film is placed on the optical table base, a gold film surface of the optical window is toward the infrared light spectrum pool, and the middle of the optical window is sealed by the PDMS sealing ring; the light spectrum pool, the PDMS sealing ring and the silicon hemisphere optical window are screwed-in and fixed through an external thread of the fixed support and an internal thread of an upper support of the optical table base so as to form the surface enhanced infrared spectrometer for DNA analysis. The testing device, provided by the invention, has the advantages of simple structure, convenience for operation and small required sample amount, and can be stably used for a long time and convenient for real-time on-line detection of infrared spectrum signals at a waveband of 1100cm<-1> to 4000cm<-1>, and high-sensitivity analysis of DNA molecules, so that DNA identification and hybridization mechanisms and kinetics research can be realized.
Owner:NANJING UNIV
Plant tissue small sample rapid preparation method
InactiveCN103234787APreparation work is flexibleFacilitate analysis workPreparing sample for investigationBiotechnologyNicotiana tabacum
The invention relates to a plant tissue small sample rapid preparation method which is used for carrying out component analysis or DNA identification of plant fresh samples. According to the invention, a plant fresh sample is directly added into a 2ml round-bottom centrifuge tube, and is subjected to a bake-drying treatment in a baking oven or a fast-drying treatment in liquid nitrogen; the sample is directly crushed in the centrifuge tube by using a handheld drill with a dome bit; an extraction liquid is added into the bake-dried sample tube for leaching, or an extraction liquid is added into the frozen sample tube for decomposing cells; a supernatant is obtained by separation in a high-speed centrifuge; and the supernatant is adopted as a component analysis or DNA identification sample. With the method provided by the invention, sample preparation work is flexible, instrument operation is eliminated, steps are simplified, certain test cost is saved, and working efficiency is improved. The method provided by the invention is suitable to be used in tobacco potassium nutrition and potassium high-efficiency genetic material potassium content determination works, and in sample preparation works before molecular biology research DNA extraction. The method facilitates laboratory sample analysis and testing works with small samples and large sample analysis amounts.
Owner:ZHENGZHOU TOBACCO RES INST OF CNTC
Individual DNA identification by short serial repeated sequential point isogenic gradient and determination reagent box
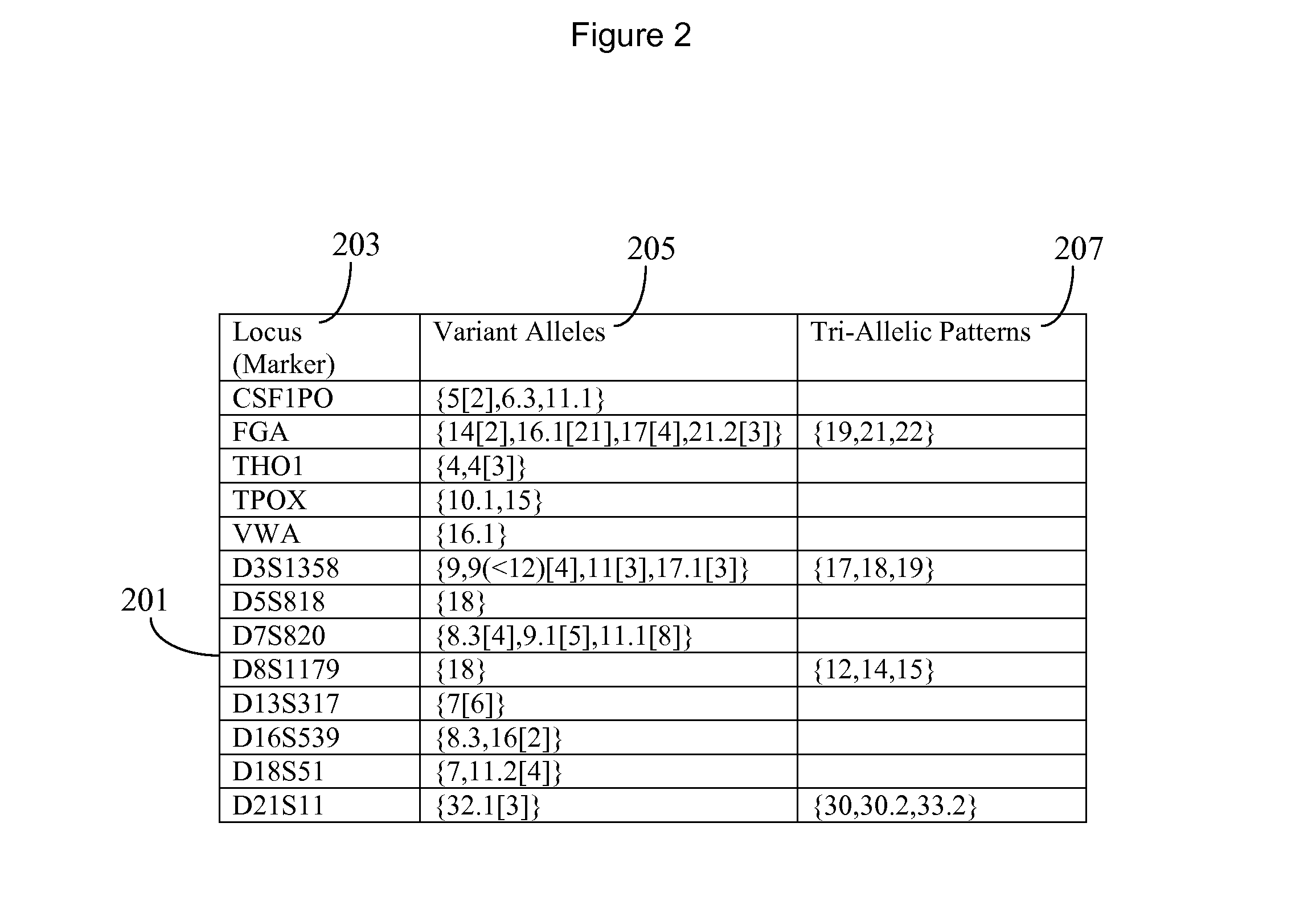
InactiveCN1644709AImprove targetingStrong specificityMicrobiological testing/measurementFluorescenceElectrophoresis
An identification method and its regent box for recognizing the unit DNA by the allelic ladder of short series-wound repeating serial locus have been opened the invention. The process is: Screening the SRT locus, designing the common reading things for synthesizing special PCR, then to mark the leading things, respectively. Then to extract the DNA of the human, going on the PCR reaction, detecting the part by electrophoresis and purifying the part. Last to connect the product of PCR to the carrier, then getting the clone of the different genes, making the every allele PCR reaction by using the leading thing of fluorescent number, mixing the PCR products of allele for STR locus then adding to the stabilizer. So we can get the allelic ladder. The ladder is the regent box to recognize the DNA. The invention needn't to collect the different gene blood again and again. And the box cost little. It is easy to make, also have good stability.
Owner:SHANGHAI SHENYOU JIANHAI BIOLOGICAL TECH LIABILITY +1
STR gene data analysis method
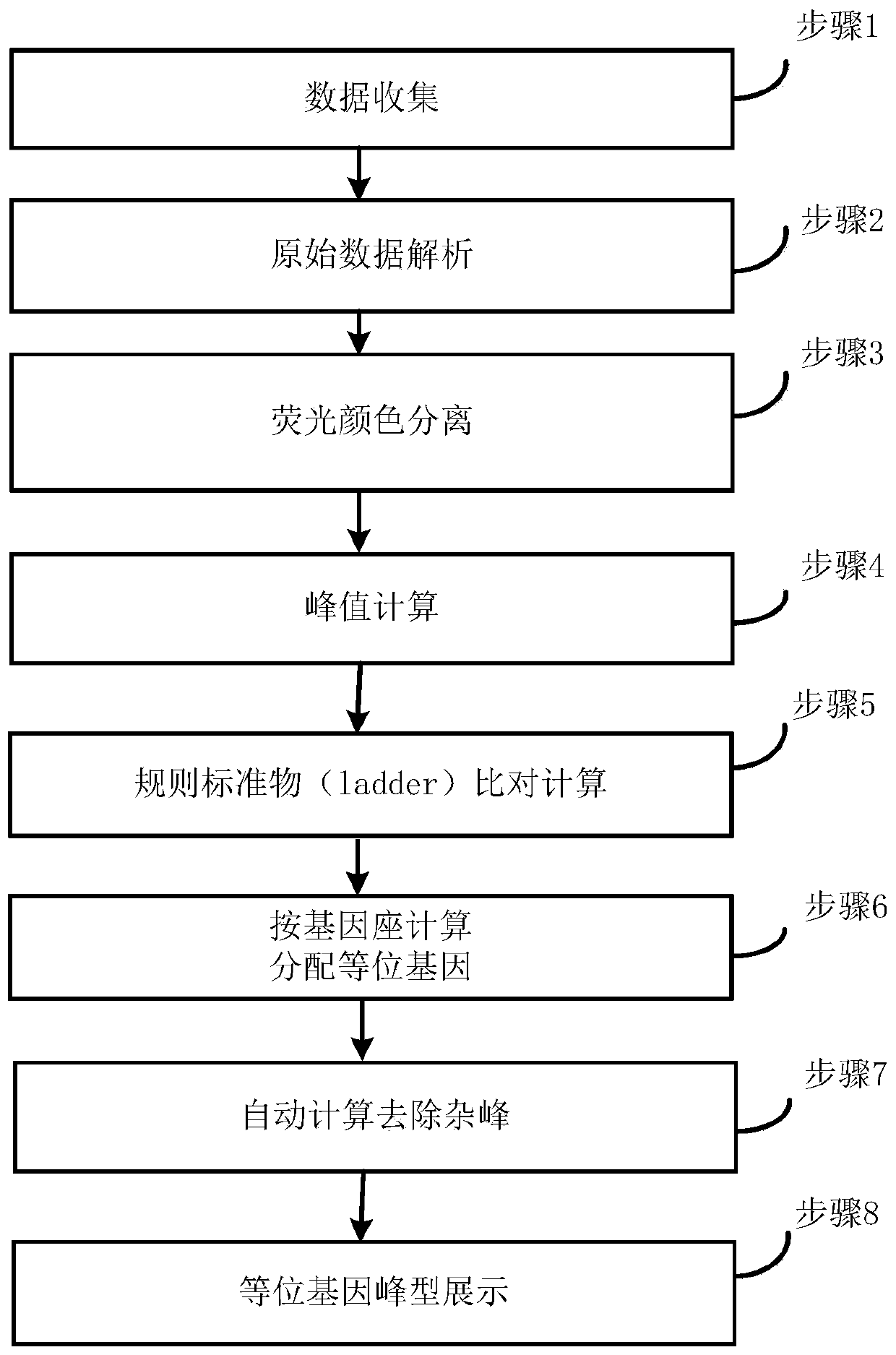
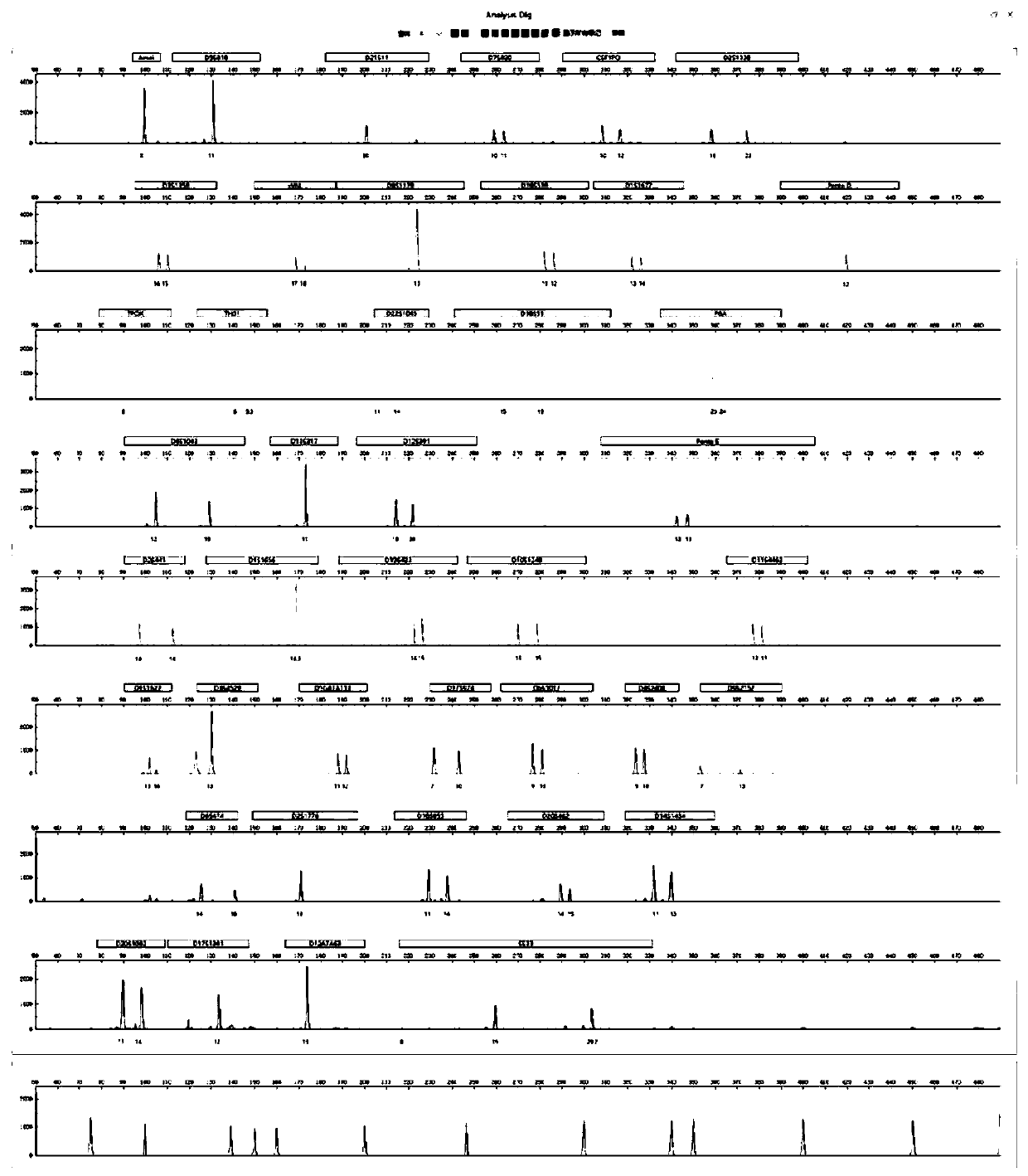
ActiveCN111415704ARealize fully automatic data analysisAccelerate the development process of localizationBiostatisticsProteomicsOriginal dataFluorescence
The invention discloses a STR gene data analysis method. On the basis of C + + language, short tandem repeat (STR) gene sequencing data in DNA is subjected to positioning full-automatic analysis, thedata processing efficiency is integrally improved, a gene region associated with specific characters is finally obtained, and a rich positioning result display chart is provided. The algorithm supports analysis of multi-color fluorescence channels, supports the kit commonly used in the market at present, suports an original data file format output by a common sequencer, can automatically identifyand remove miscellaneous peaks generated in inspection, and can automatically calibrate peak pattern dislocation generated in the sequencing process. According to the invention, full-automatic data analysis of a multi-color fluorescence color channel is carried out on a DNA gene short tandem sequence STR in DNA identification and detection by a forensic medicine. The situation that DNA sequencingonly depends on foreign software analysis is broken through, a process is established for developing domestic sequencers and domestic reagents in China, and a domestication development process in thefield of forensic identification and detection of DNA in China is accelerated.
Owner:北京博安智联科技有限公司
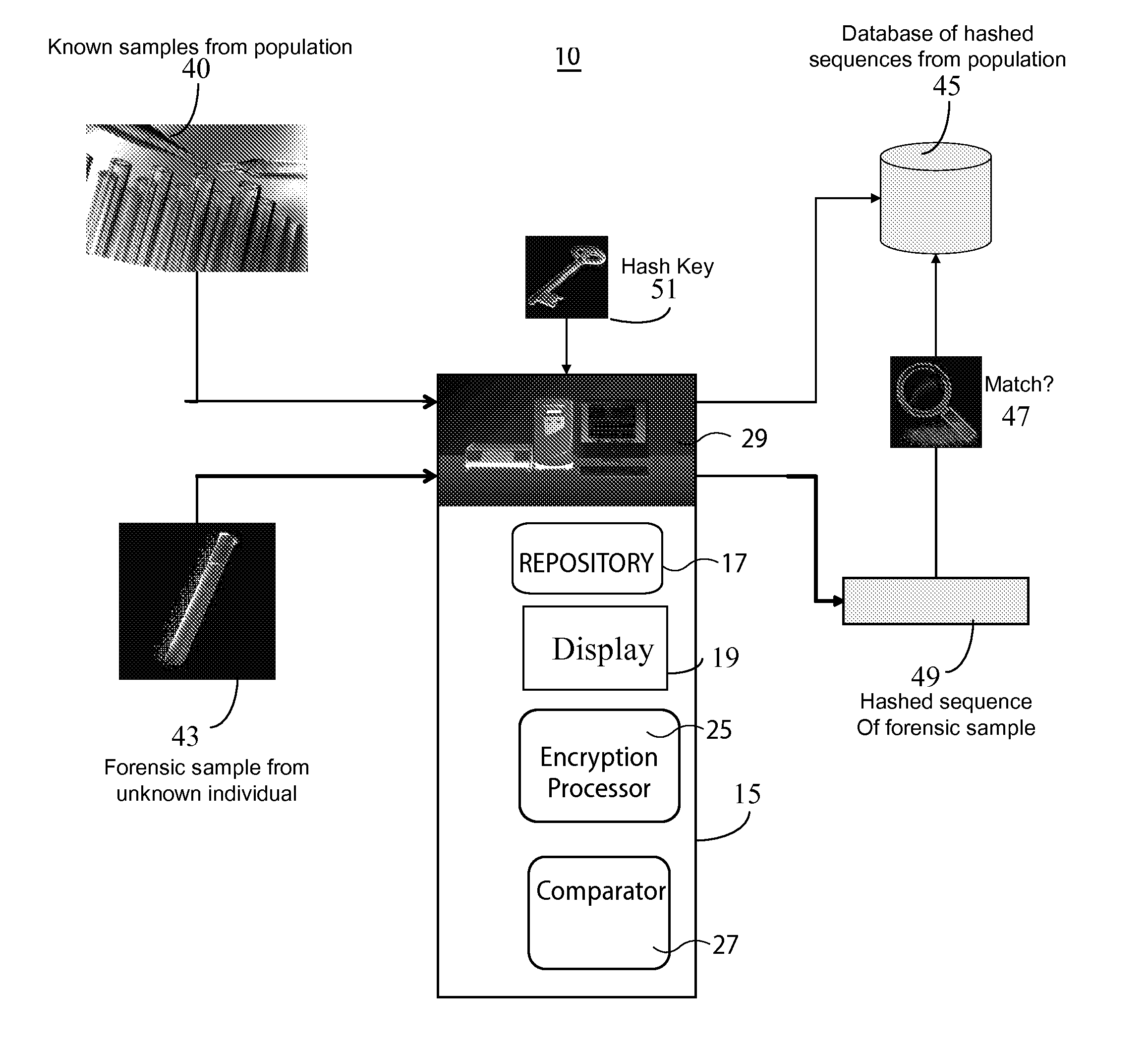
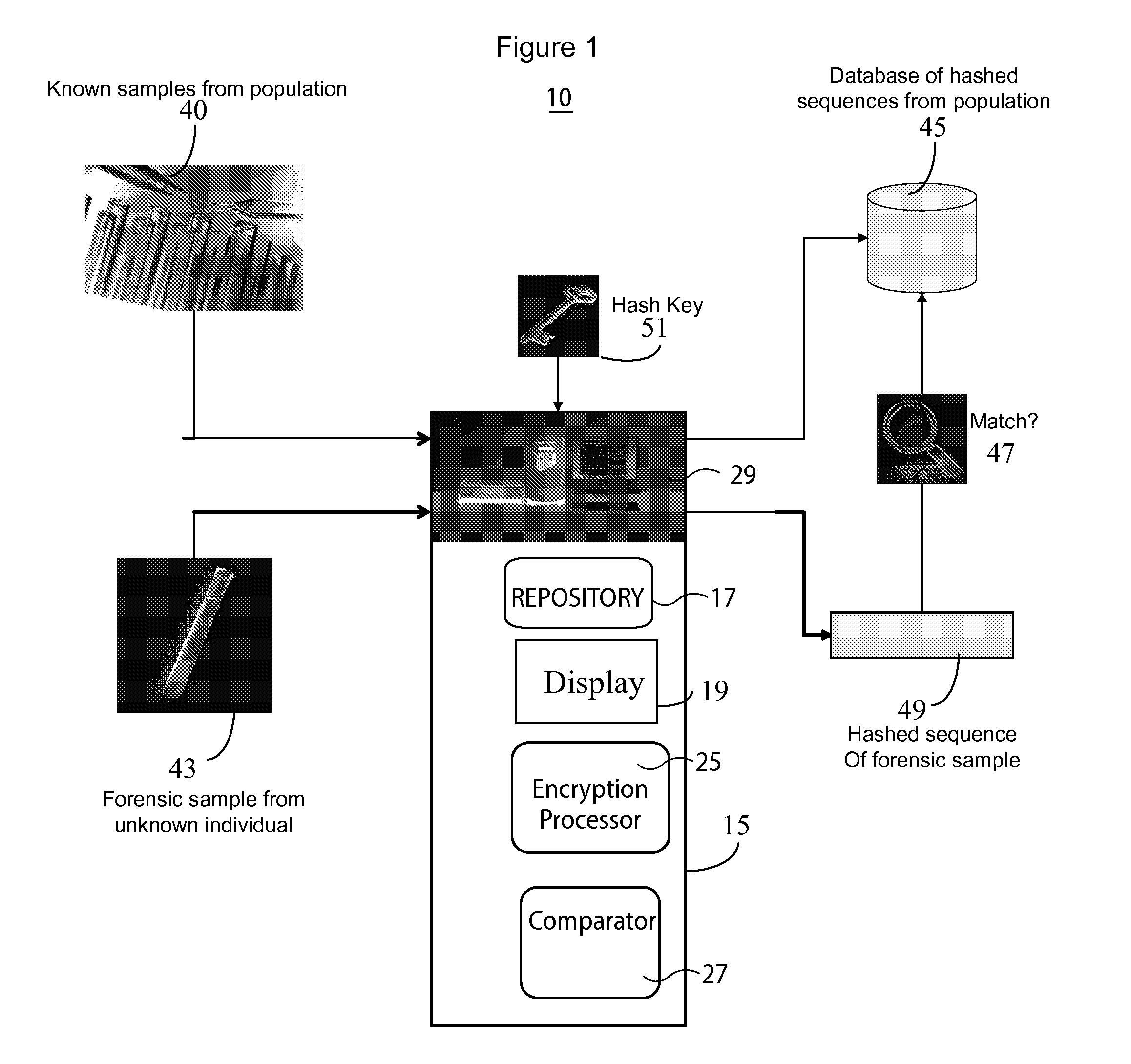
System for DNA Identification Hiding Personal Traits
InactiveUS20130266135A1Microbiological testing/measurementComputer security arrangementsGeneticsDNA sequencer
A system for DNA sequence identification hides personal and medical characteristics. A DNA sequencer processes a biological sample to provide genetic data identifying biological sample genetic marker variations of multiple different markers from corresponding reference markers. An encoding processor one way encrypts the genetic data into an encrypted code using an encryption key. A comparator compares the encrypted code with multiple encrypted codes retrieved from storage to identify a match and biological sample source. The multiple encrypted codes are derived by encrypting genetic data of multiple different biological samples using the encryption key and the multiple different biological samples are associated with corresponding identifiers of their respective biological sample sources.
Owner:CERNER INNOVATION
Multiplex DNA identification of clinical yeasts using flow cytometry
InactiveUS7465543B1Readily apparentMicroorganismsMicrobiological testing/measurementAscomycetous yeastPathogenic yeast
Oligonucleotide probes that are specific for pathogenic yeasts and which may be used for the detection of such yeasts in biological samples are described. These probes are specific for yeast species within the clade of Candida albicans and other pathogenic ascomycetous yeasts, and may be used singly or in a multiplex hybridization assay system.
Owner:UNITED STATES OF AMERICA AS REPRESENTED BY THE SEC OF AGRI THE
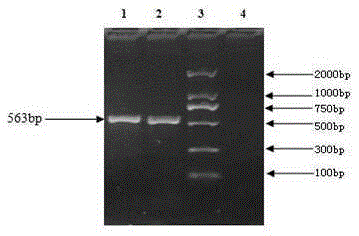
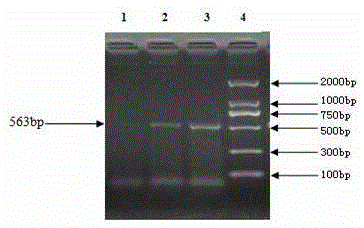
Deer fetus DNA identification kit and method
InactiveCN105238856AThe identification method is simpleRapid identification methodMicrobiological testing/measurementBiotechnologyDNA extraction
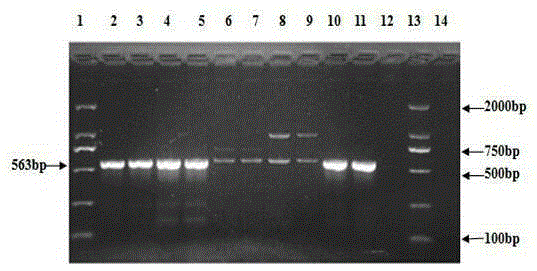
The invention relates to the technical field of traditional Chinese medicine detection, in particular to a deer fetus DNA identification kit and method. The deer fetus DNA identification kit comprises a mitochondria DNA extraction system, a PCR (polymerase chain reaction) system and a result observation system. The deer fetus DNA identification method includes the steps of detection sample pretreatment, mitochondria DNA extraction, PCR primer designing and synthesis, building of the PCR system and result judgment, wherein judgment standards includes that deer fetuses are judged to be qualified if 563bp strips appear and judged to be pseudo if only multiple strips or no strips appear. The PCR identification method has the advantages of simplicity, quickness and reliable detection result, and specificity of qualified and pseudo deer fetuses can be accurately differentiated and identified at the same time.
Owner:BEIHUA UNIV
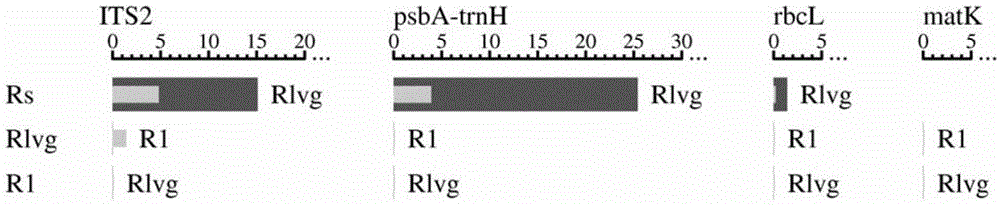
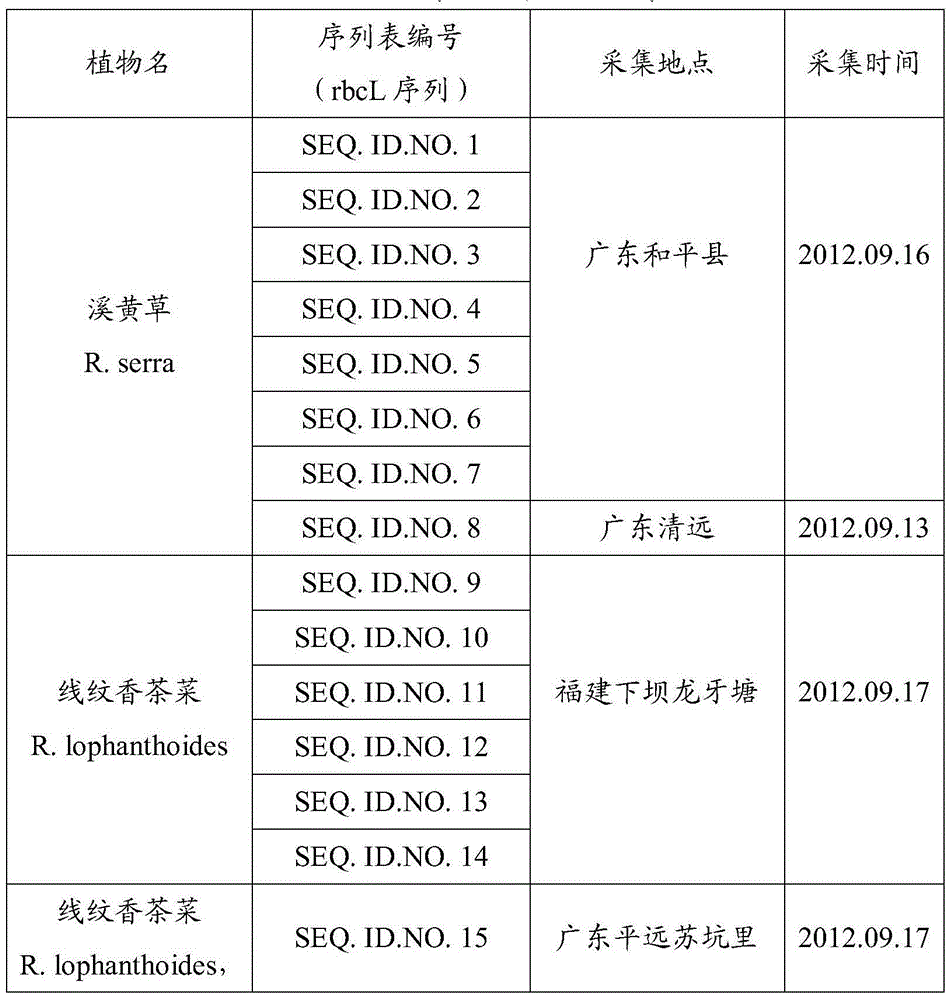
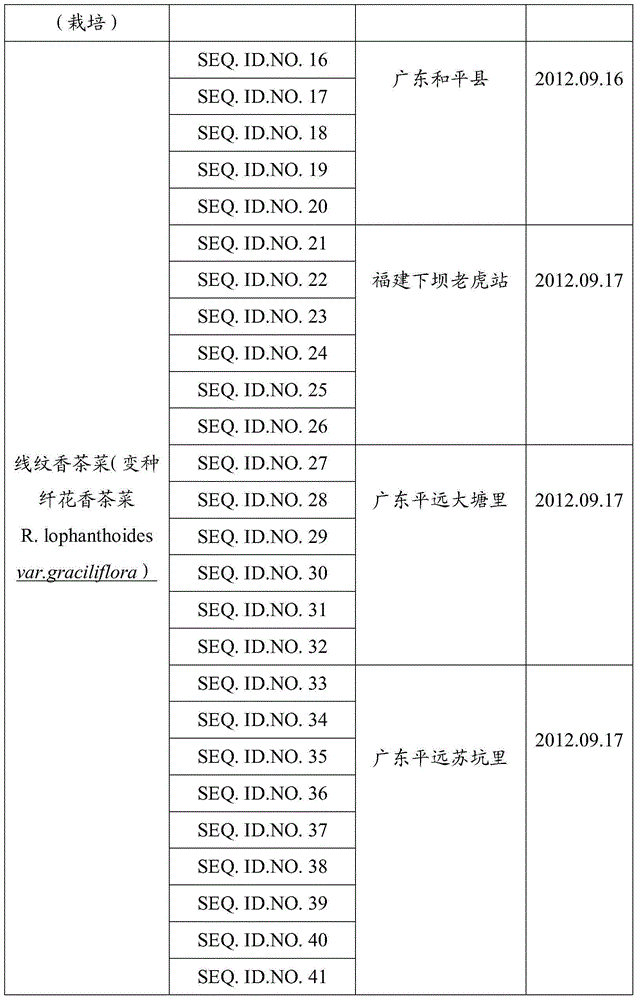
DNA identification method of Isodon serra(Maxim.)Kudo and Rabdosia lophanthoides (Buch.-Ham. ex D. Don) Hara var. graciliflora (Benth.) Hara
ActiveCN104404629AEasy to expandEasy to compareMicrobiological testing/measurementProtein nucleotide librariesLamiaceaeGene
The invention provides a gene pool containing rbcL gene sequence fragments of labiate Isodon serra(Maxim.)Kudo and Rabdosia lophanthoides (Buch.-Ham. ex D. Don) Hara var. graciliflora (Benth.) Hara. The invention also provides a method for identifying plant species of the labiate Isodon serra(Maxim.)Kudo and Rabdosia lophanthoides (Buch.-Ham. ex D. Don) Hara var. graciliflora (Benth.) Hara. The method comprises the step of comparing a rbcL gene or its sequence fragment of a plant sample, plant species of which is to be identified, to the gene pool provided by the invention. The invention also provides an application of the rbcL gene or its sequence fragment in identifying the labiate Rabdosia lophanthoides (Buch.-Ham. ex D. Don) Hara var. graciliflora (Benth.) Hara and the labiate Isodon serra(Maxim.)Kudo. It proves that the rbcL sequence is general, is easy to amplify and compare, and has good amplification and identification effects on the labiate Rabdosia lophanthoides (Buch.-Ham. ex D. Don) Hara var. graciliflora (Benth.) Hara and the labiate Isodon serra(Maxim.)Kudo.
Owner:GUANGZHOU BAIYUSN HUTCHISON WHAMPOA CHINESE MEDICINE
Method for screening DNA bar code universal sequences of seven major forage grass varieties of gramineous family
InactiveCN106047999AImprove recognition rateMicrobiological testing/measurementAnimal ForagingConserved sequence
The invention discloses a method for screening NA bar code universal sequences of seven major forage grass varieties of gramineous family. The method comprises the following steps: (1) firstly, in accordance with nucleotide sequences of gramineous forage grass matk and rbcl genes in GenBank, designing four pairs of universal primers, establishing amplification conditions for target segments of 11 samples of eight forage grasses of seven major forage grass varieties of the gramineous family and conducting amplification; (2) sequencing and analyzing an amplification product, and conducting comparison, screening out 5'-end and 3'-end conserved sequences of four marker sites of the seven major forage grass varieties, and conducting single nucleotide polymorphism haplotype analysis on nucleotide within the conserved regions of the various marker sites, knowing that matk1 has six haplotypes, matk2 has seven haplotypes, matk3 has three haplotypes and the rbcl gene has five haplotypes; and (3) in accordance with four marker points screened from the matk and rbcl genes, establishing specific DNA identification codes corresponding to the eight forage grasses. The method disclosed by the invention is good in primer university and ideal in amplification effect, and meanwhile, the method is high in species identification rate.
Owner:GANSU AGRI UNIV
Reliable and Secure Detection Techniques for Processing Genome Data in Next Generation Sequencing (NGS)
Genetic samples are obtained from separate people, and at least a portion of each are purposefully combined before testing to form a pooled genetic sample. The pooled genetic sample is tested for the presence of a signature for a given known ailment. DNA identification uses discovered InDels in a region of InDel variation in a genetic sample. A pair-wise comparison is performed to reference InDels, and a distance is measured between the first InDel and the reference Indel. Reference kmers are identified in a reference genome, and in a test sample. The plurality of sample kmers are filtered to those which have a 1 edit distance from a corresponding one of the plurality of reference kmers. Reads that have kmers that do not have a 1 edit distance from the corresponding one of the plurality of reference kmers are identified, and multiple single-mutations are eliminated from candidate InDel reads.
Owner:CRYSTAL GENETICS INC
Benzothiazole and triazolediheterocycle-containing fused ring compound and application thereof
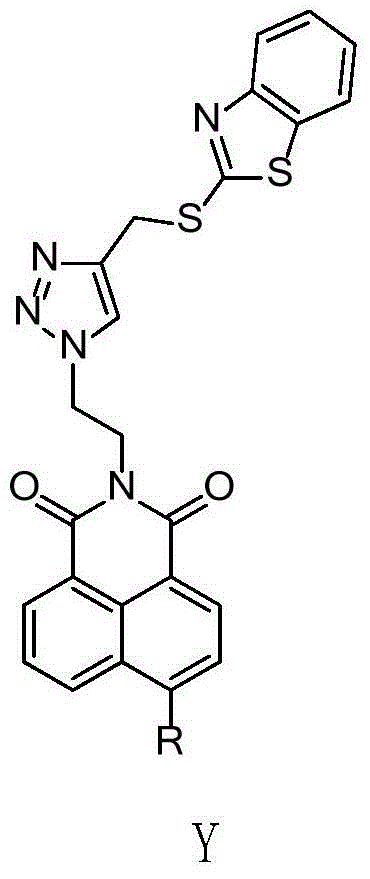
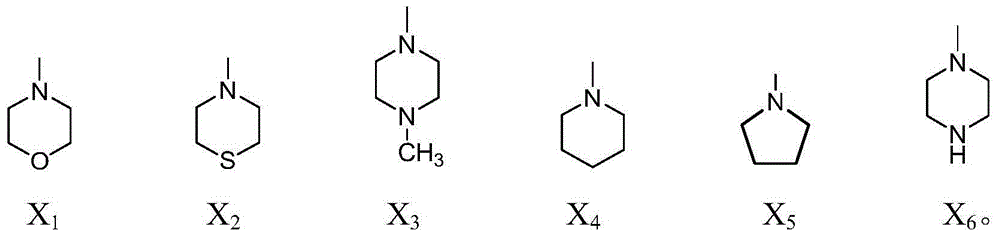
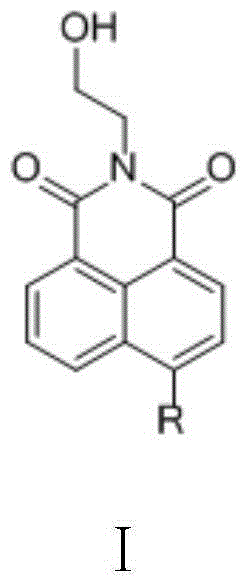
InactiveCN104059062AImprove biological activityIncrease diversityOrganic chemistryAntineoplastic agentsSide chainNormal growth
The invention relates to a benzothiazole and triazolediheterocycle-containing fused ring compound which has a structure of a general formula Y shown in the specification. According to the compound, cyclic amine with bioactivity is connected to the site 4 of naphthalimide, a structure that an imine nitrogen atom of naphthalimide is spaced from a triazole nitrogen atom connected with a side chain by two carbon atoms is kept, and molecules are introduced into sulfur-containing heterocyclic benzothiazole through linkage of triazole, so that the diversity of naphthalimide derivatives, the bioactivity of naphthalimide and the DNA identification and intercalation capability are improved; the compound has a relatively good inhibition effect on normal growth of tumor cells, with different tissue sources, of cervical cancers, liver cancers, breast cancers and the like.
Owner:DALIAN UNIV OF TECH
Standard black sea bream DNA bar code detecting sequence and application thereof
InactiveCN107034290AAccurate identificationOvercoming the inability to identifyMicrobiological testing/measurementVector-based foreign material introductionDNA barcodingBlack sea
The invention discloses a standard black sea bream DNA bar code detecting sequence and application thereof. The sequence is shown as SEQ ID NO:1. By adopting a black sea bream molecule identification method, a reliable DNA identification technology adopted for individuals which are incomplete or cannot accurately identified in specimens and complete specimens is achieved, and the technical defect that in the prior art, incomplete individuals cannot be identified and complete individuals can be identified by needing specialized persons very familiar with the morphology of black sea breams is overcome. The black sea breams can be rapidly and accurately identified compared with a traditional morphological identification method.
Owner:ZHENJIANG HUADA DETECTION CO LTD
Genetic marker group and individual gene identification card and application thereof
InactiveCN109321660AImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationIndividual geneHaplotype block
The invention discloses a genetic marker group and an individual gene identification card and application thereof. The genetic marker group comprises multiple SNP sites, wherein the multiple SNP sitesare tagSNPs located in different haplotype areas, the MAF is between 0.48 to 0.505, the adjacent SNP sites are not in linkage relationships, the distance among the adjacent SNP sites is larger than 1M, and variation information non A / T or C / G and the SNPs are evenly distributed in all chromosomes. By applying the technical scheme, through effective combination of the SNP sites, the set of genetic marker group which is simple in technology, economical and practical is formed, and can be used as the individual DNA identification card, and when the SNP sites are selected, linkage relationships,the MAF, distance, variation situations and the evenness of distribution on the chromosomes of the sites are taken into account, so that the accuracy of judging a certain gene sample is improved.
Owner:BEIJING USCI MEDICAL LAB CO LTD
Manufacturing method for hair handicraft picture
InactiveCN102837544AStrong adhesionWon't fall offSpecial ornamental structuresCooking & bakingEngineering
The invention provides a manufacturing method for a hair handicraft picture, which comprises the following steps: A) manufacturing a background picture; B) collecting hairs; C) treating the hairs: disinfecting and dyeing the hairs collected in the step B); D) sticking: sticking the hairs to the background picture by using glue; and E) baking: putting the handicraft picture manufactured in the step D) into an oven for heating and baking. According to the manufacturing method for the hair handicraft picture provided by the invention, the hairs are remained on the picture, so that the permanent memento can be kept and can be inherited for lifetime; the significance is deep; the manufacturing method is special; the hairs are firmly stuck to the picture and do not fall off after a long time; the color is not changed; the hairs of a user or a relative are remained on the picture, so that a material for the future DNA identification is supplied; and the existing hairs are used for manufacturing, so that the cost is saved.
Owner:李俊江
Polyamide bisbenzimidazole compound, preparation method and application thereof
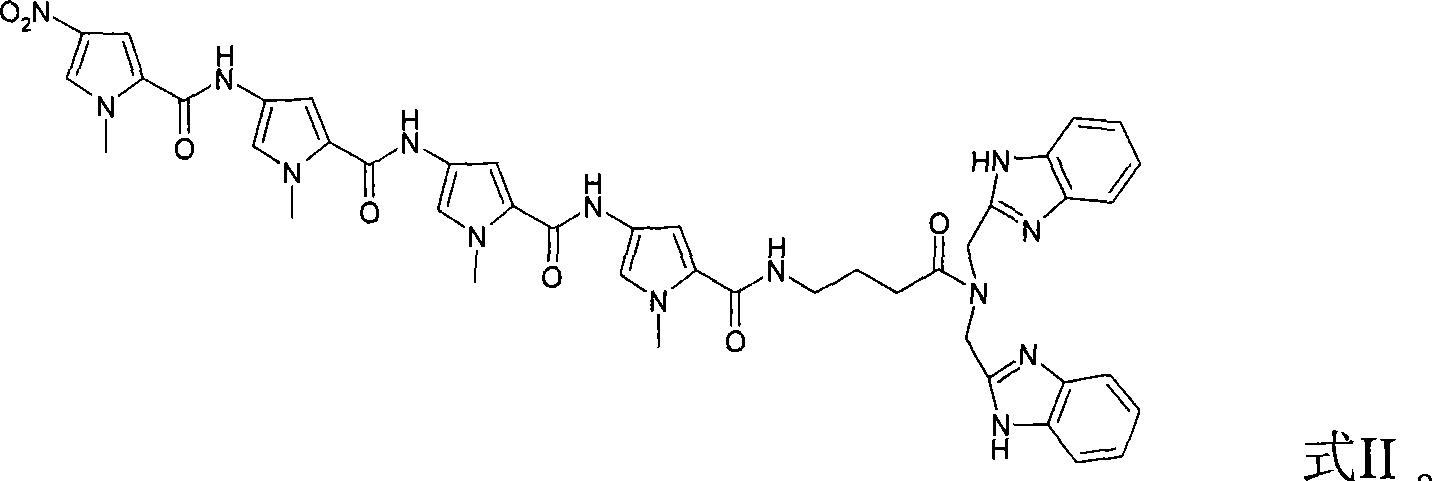
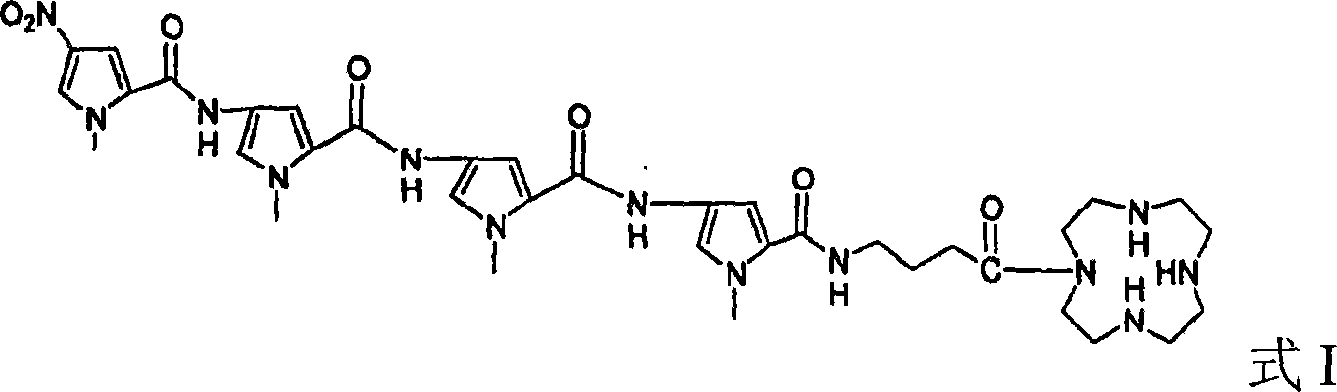
InactiveCN101085773AShort reaction timeLower reaction costSugar derivativesDNA preparationPolymer scienceAcyl group
The invention relates to a kind of polyamide benzimidazole compound and the preparing method and its application. The structure II of said compound is as follows. The preparation method comprises following steps: preparing 4- (4- (4- (4- nitro- N- methyl pyrrole- 2- acyl)ammino- N- methyl pyrrole- 2- acyl)ammino- N- methyl pyrrole-2- acyl) ammino- N- methyl pyrrole- 2- acyl)ammino butyrate, adding 1- hydroxy benzotriazole and dicyclo hexylcar bodiimide with its mole being 2- 4 times of that of said product, reacting at 20- 30 Deg. C for 4- 12 hours, adding (2- benzimidazole methylene) amine, reacting at 20- 30 Deg. C for 4- 16 hours, and getting target product. The invention aslo provides nucleic acid cutting agent taking the metallic complex as active component of said compound. The invention provides good base and effect way for further synthesis of DNA identification and cutting agent.
Owner:BEIJING UNIV OF CHEM TECH
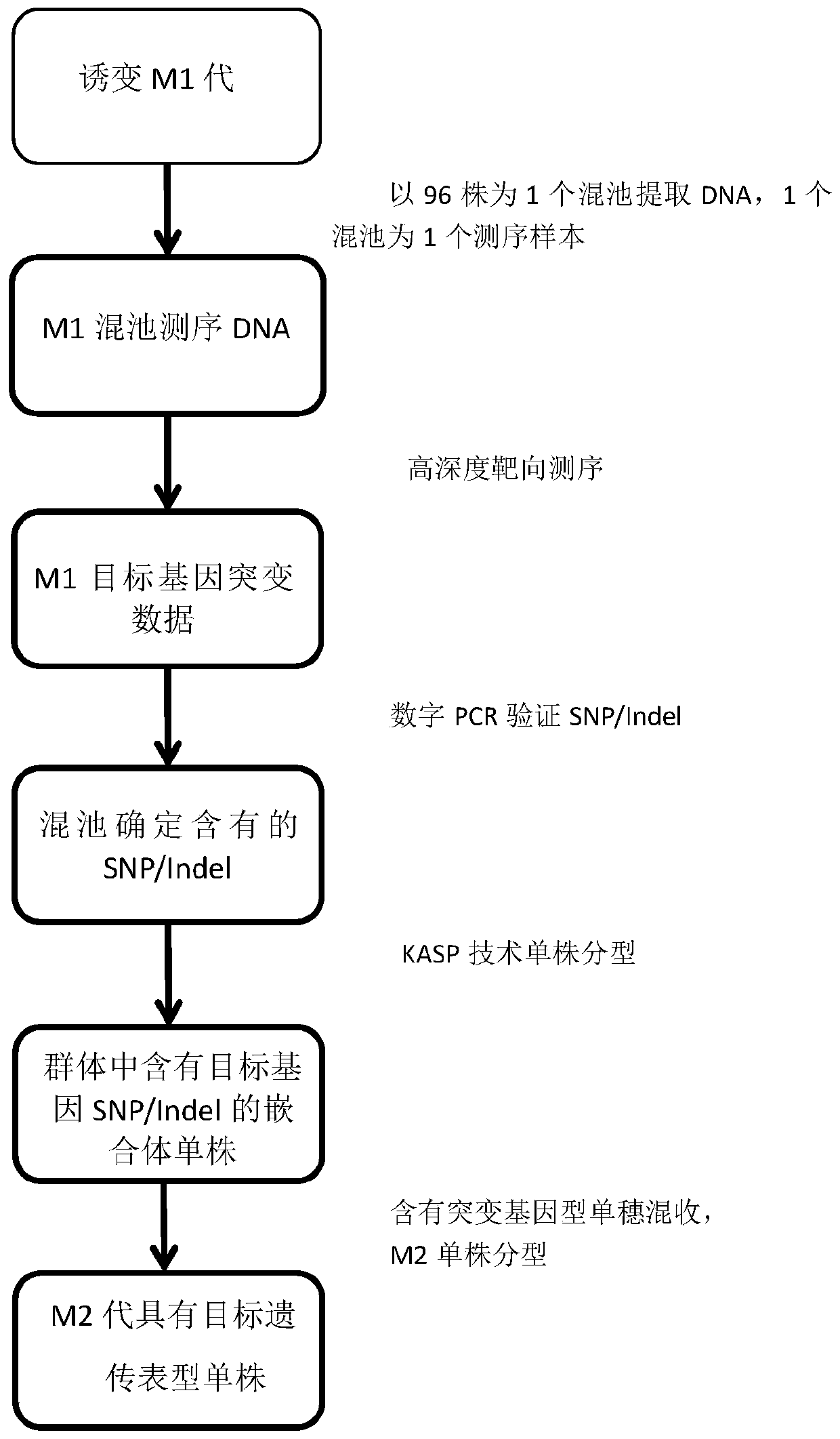
Method for high-throughput targeted identification of M1-generation mutation of physicochemical mutated plant and obtaining of mutant
ActiveCN110904258ASave time and costMicrobiological testing/measurementPlant genotype modificationBiotechnologyGermplasm
The invention discloses a method for high-throughput targeted identification of M1-generation mutation of a physicochemical mutated plant and obtaining of a mutant. The identification method comprisesthe following steps: mutagenizing a plant in a non-lethal dosage physicochemical mutagenesis mode, planting the obtained M1-generation plant into single plants, mixing the leaves of each single plant, extracting polled DNA, and performing high-depth targeted sequencing on a target gene region; and comparing the obtained sequencing result with the related sequence of a target gene region, and identifying whether the target SNP and / or Indel exists or not. DNA identification of the chimeric single plants is carried out on the basis of identifying the chimeric single plants containing target generegion mutation, selecting ears containing the mutation, harvesting seeds of the ears in a mixed mode, performing mixed sowing, and then carrying out DNA identification on the single plants to obtainan M2 single plant with a target genetic phenotype. The method has the advantages of high efficiency, good accuracy, and simplicity in operation, and is of remarkable progressive significance for identification and acquisition of innovative germplasm.
Owner:HUNAN HYBRID RICE RES CENT
Chinese medicine DNA detection kit and identification method
InactiveCN101434989AEasy to useThe identification method is simpleMicrobiological testing/measurementPenisPositive control
The invention relates to the Chinese medicine detection technology field and belongs to a Chinese medicine DNA detection kit, comprising an identification reaction system, a positive control reaction system and a negative control reaction system; both the identification reaction system and the positive control reaction system contain buffer solution, 12.5mM dNTP, one type of 0.1mM deer penis primer, fritillary primer and root of fangji primer, one type of Taq DNA polymerase, deer penis, fritillary and root of fangji DNA, sample DNA to be detected, and double-distilled water; the negative control reaction system contains buffer solution, 12.5mM dNTP, one type of 0.1mM deer penis primer, fritillary primer and root of fangji primer, one type of Taq DNA polymerase, bullwhack, false fritillary and fake root of fangji DNA, and double-distilled water. The Chinese medicine DNA identification method comprises steps such as designing deer penis, fritillary, root of fangji mitochondrial DNA three-pair specific oligonucleotide primer, artificial synthesis, fixing reaction procedures and result judgment, and the like; the Chinese medicine DNA identification kit can accurately identify the specificity of Chinese medicinal materials; and the identification method has the advantages of simplicity, rapidness, and reliable detection result, etc.
Owner:BEIHUA UNIV
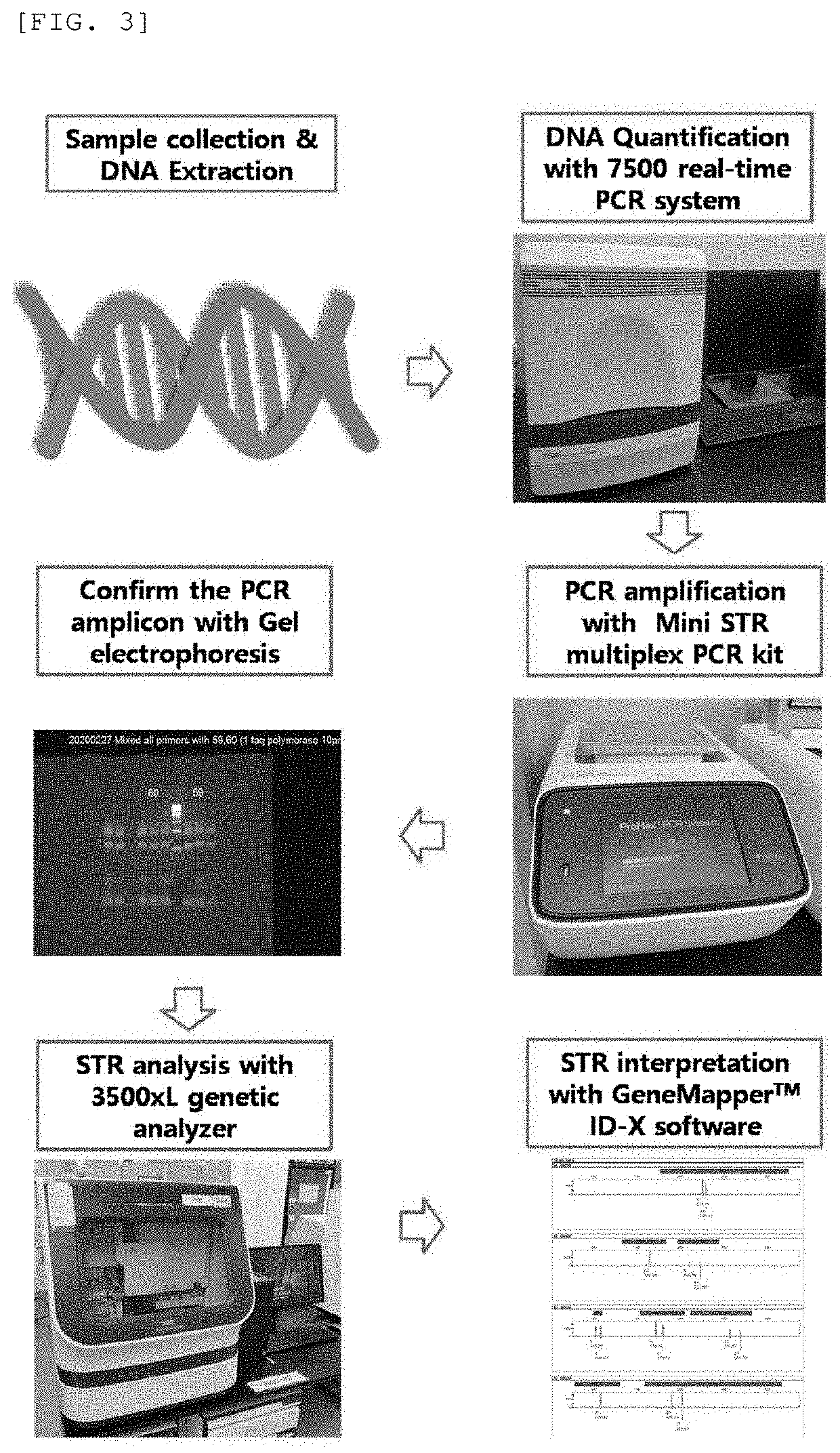
Multiplex PCR Kit for identifying human genotype profile using new combination of mini STRs and method for identifying human genotype profile using the same
PendingUS20220028484A1Improving precision in identity determinationIncrease success rateMicrobiological testing/measurementProteomicsMultiplexMedicine
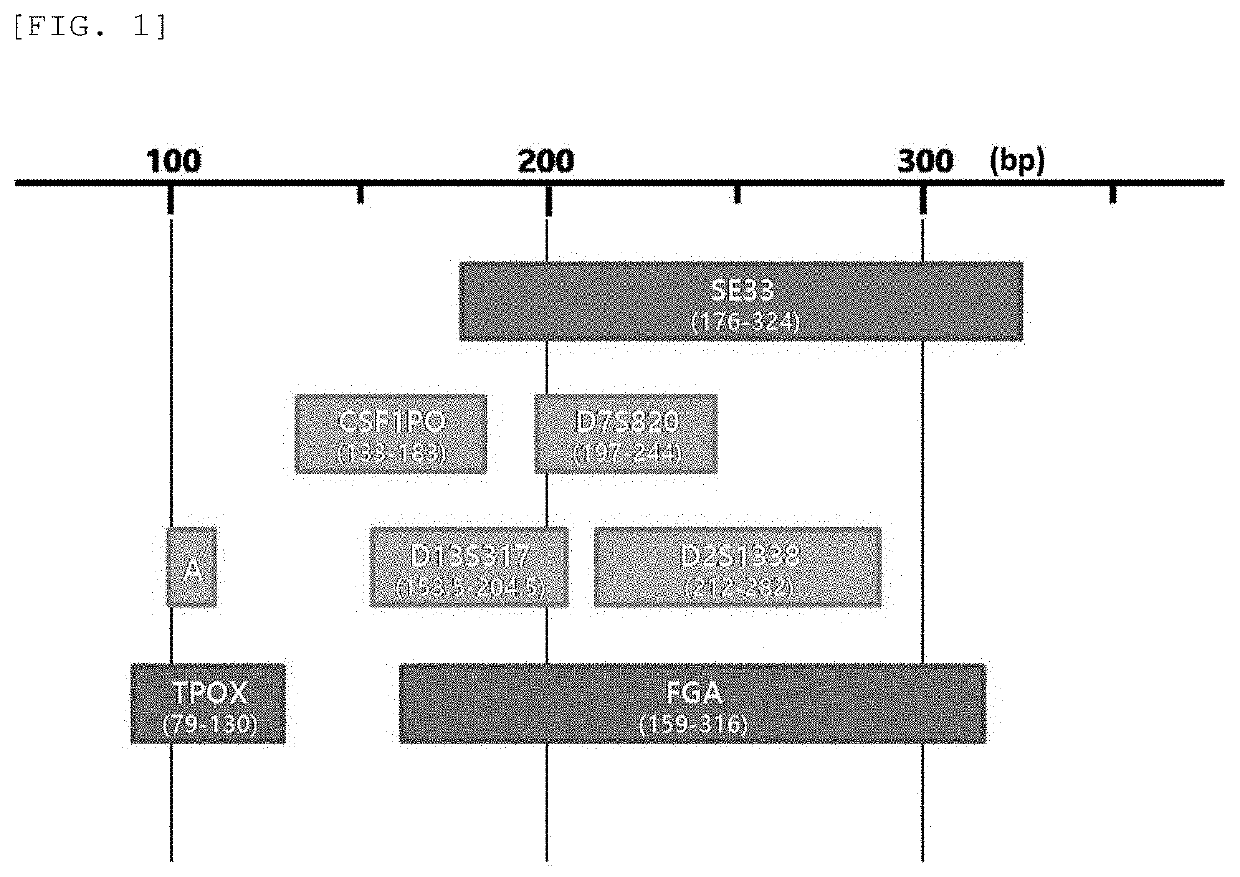
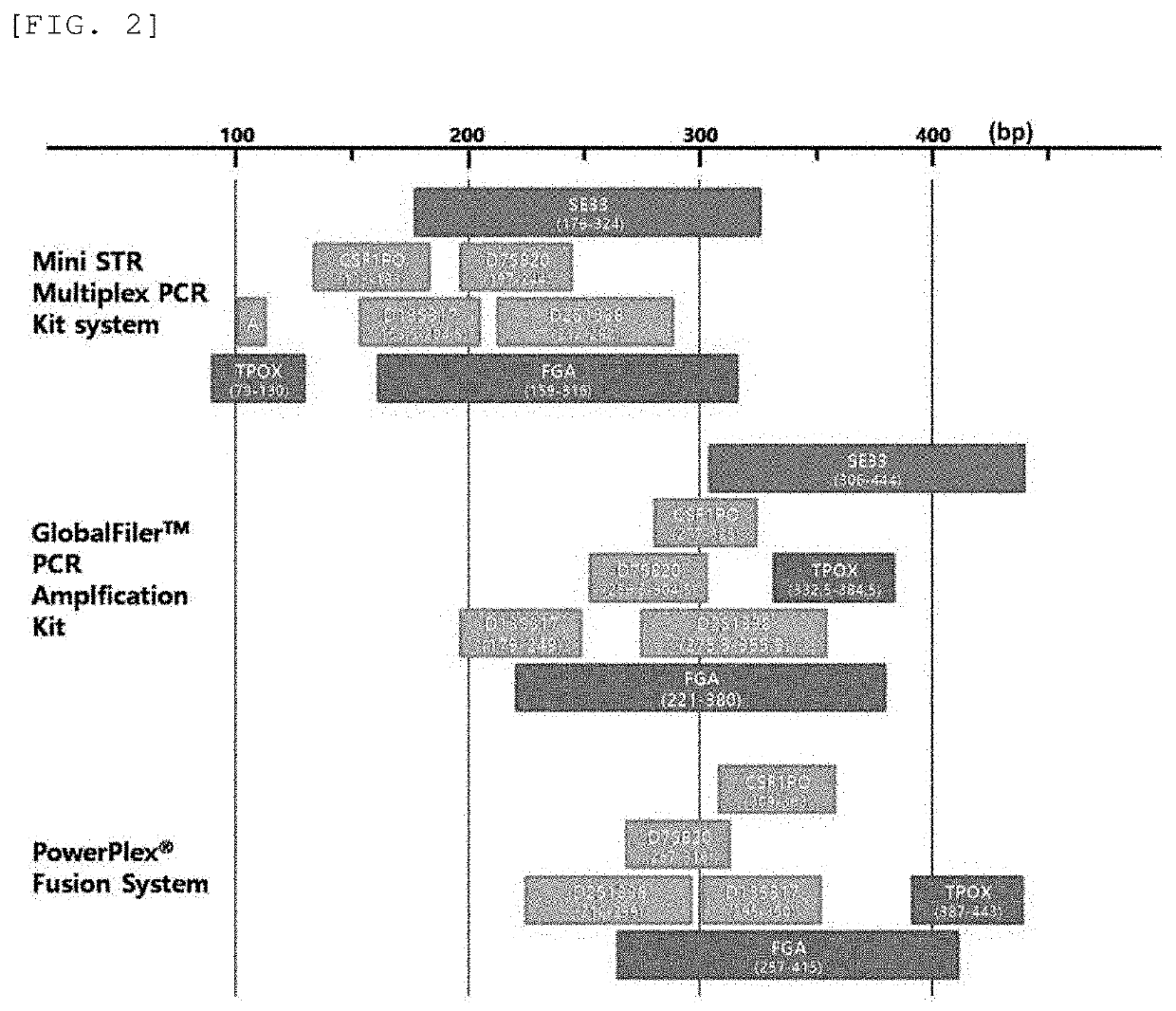
The present invention provides a multiplex PCR kit with improved STR analysis success rate by introducing mini-STRs at 7 gene loci including a mini-STR at SE33 locus, which has high gene identification sensitivity, into a human genotype profile identification technique, as well as a method for identification of human genotype profiles using the same. According to the present invention, the STR analysis success rate is improved by identifying an STR at a locus that cannot be discriminated depending on DNA degradation level, thereby offering a lot of DNA information. Further, the present invention may contribute to more efficient DNA identification such as extension of database record, increase in identity determination success rate, etc. Still further, clues for reconstruction of a crime scene may be offered while improving reliability of forensic investigation results, simultaneously, thereby contributing to solving the crime.
Owner:REPUBLIC OF KOREA (NTL FORENSIC SERVICE DIRECTOR MINIST OF PUBLIC ADMINISTRATION & SECURITY)
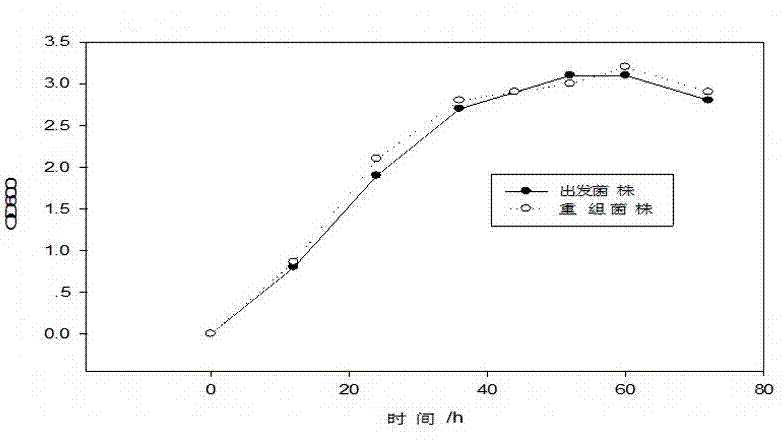
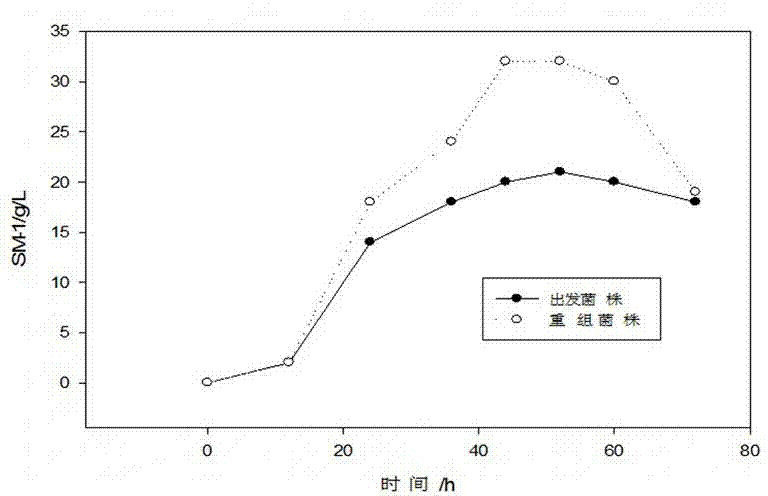
High-specific heat capacity microbe biological polysaccharide SM-1 and its preparation method and use
InactiveCN104498561AReduce elevationReduce churnCosmetic preparationsOrganic active ingredientsSide chainFermentation
The invention belongs to the technical field of microbe polysaccharide and relates to high-specific heat capacity microbe biological polysaccharide and its preparation method and use. The preparation method comprises that a bacillus subtilis strain capable of effectively accumulating extracellular capsular polysaccharide is selected from nature and then is subjected to 16rs DNA identification, a polysaccharide synthesis approach-related gene is adjusted and modified so that a yield is improved, and through microbial fermentation, alcohol precipitation, and drying and refining, the high-specific heat capacity microbe biological polysaccharide is prepared. The high-specific heat capacity microbe biological polysaccharide has a structure skeleton mainly comprising glucose, galactose, mannose and glucuronic acid, has a side chain tail end obtained by connection polymerization of pyruvic acid and acetyl groups, is a polymer, has molecular weight of 5000-30000Da, is named as SM-1, has a good water retention capacity and a high-specific heat capacity, has moisture retention and infrared radiation resistance, and has a good application value in the cosmetic industry.
Owner:杨红兵
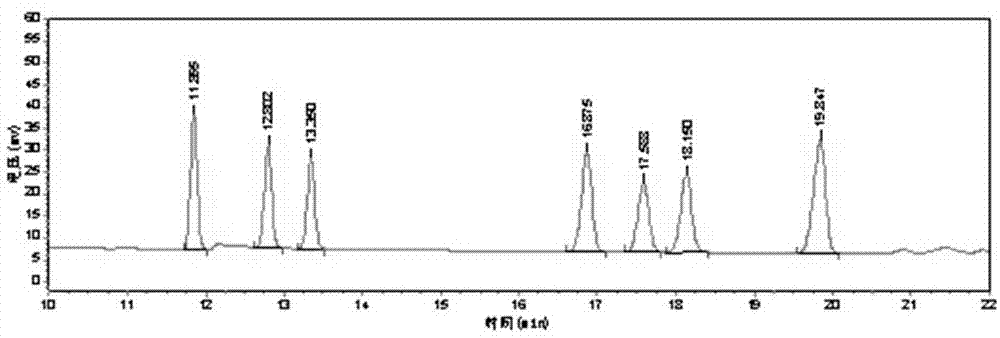
Zaocys dhumnades capillary electrophoresis DNA fingerprint spectrum and identification method
InactiveCN107699614AA large amountHigh polymorphismMicrobiological testing/measurementCapillary electrophoresisFluorescence
The invention relates to a traditional Chinese medicine zaocys dhumnades DNA identification technology, in particular to an SSR fluorescence labeled zaocys dhumnades capillary electrophoresis DNA fingerprint spectrum and identification method. The method has the characteristics of being high in resolution, convenient to operate, quick, capable of obtaining accurate results and the like and can beused for identifying zaocys dhumnades samples.
Owner:BEIHUA UNIV
Method for breeding low temperature-resistant large yellow croakers
InactiveCN101606502BGood resistance to low temperature environmentClimate change adaptationPisciculture and aquariaAquatic productSeawater
The invention belongs to the field of seawater aquaculture and particularly relates to a method for breeding and identifying low temperature-resistant large yellow croakers (also known as Pseudosciaena crocea). The invention mainly solves the status quo that a majority of large yellow croakers are not capable of accommodating to low-temperature culture currently; and the invention provides a method for breeding low temperature-resistant large yellow croakers by selecting large yellow croaker individuals capable of naturally resisting the low temperature and breeding on preferential basis, andprovides a method for identifying low temperature-resistant large yellow croakers at the same time. The low temperature-resistant large yellow croakers are obtained through multi-generation breeding and culturing, observation-based identification and DNA identification.
Owner:ZHEJIANG OCEAN UNIV
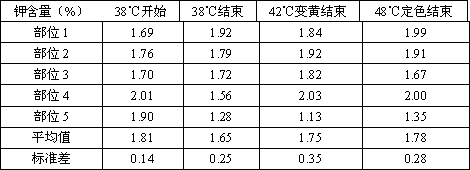
Method for preparing tobacco-leaf-tissue PCR template
The invention relates to a method for preparing a PCR template, in particular to a method for preparing a tobacco-leaf-tissue PCR template. The method includes the steps that a NaOH solution is an extracting liquid, the extracting liquid and plant leaf tissue are put into a centrifuge tube, the mixture is directly subjected to sample grinding on a sample grinding machine, and the obtained supernatant liquid directly serves as the template to carry out a PCR reaction. The method has the advantages that the heating step, the boiling step, the neutralizing step and the like are not required, the supernatant liquid can be directly used for the PCR reaction template; the prepared DNA template PCR amplification result is stable, accurate and good in repeatability, and compared with the DNA obtained with the traditional SDS extracting method and the kit extracting method, the PCR result is free of obvious difference; the method is also applied to plants such as wheat, sorghum, rice and corn; devices used for the method are small, sample consumption is small, the method is rapid, simple and convenient, low in cost and free of cross pollution, a large quantity of samples can be treated in the short time, and therefore a large quantity of transgenic tobacco samples can be subjected to DNA rapid extracting and DNA identification.
Owner:TOBACCO RES INST CHIN AGRI SCI ACAD
Identification method of periplaneta americana
ActiveCN110672737AAccurate identificationRapid identificationComponent separationBiological testingPeriplanetaPeptide ions
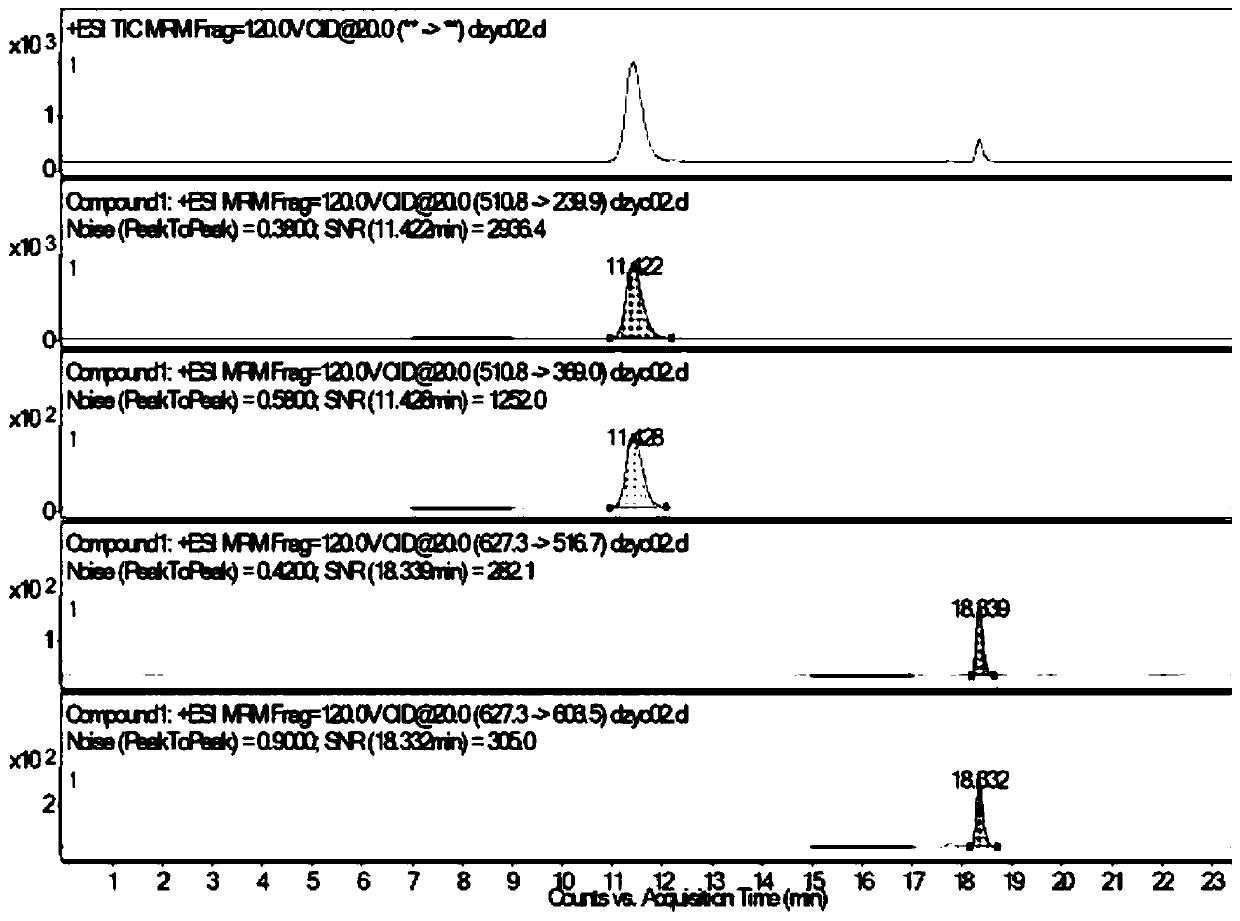
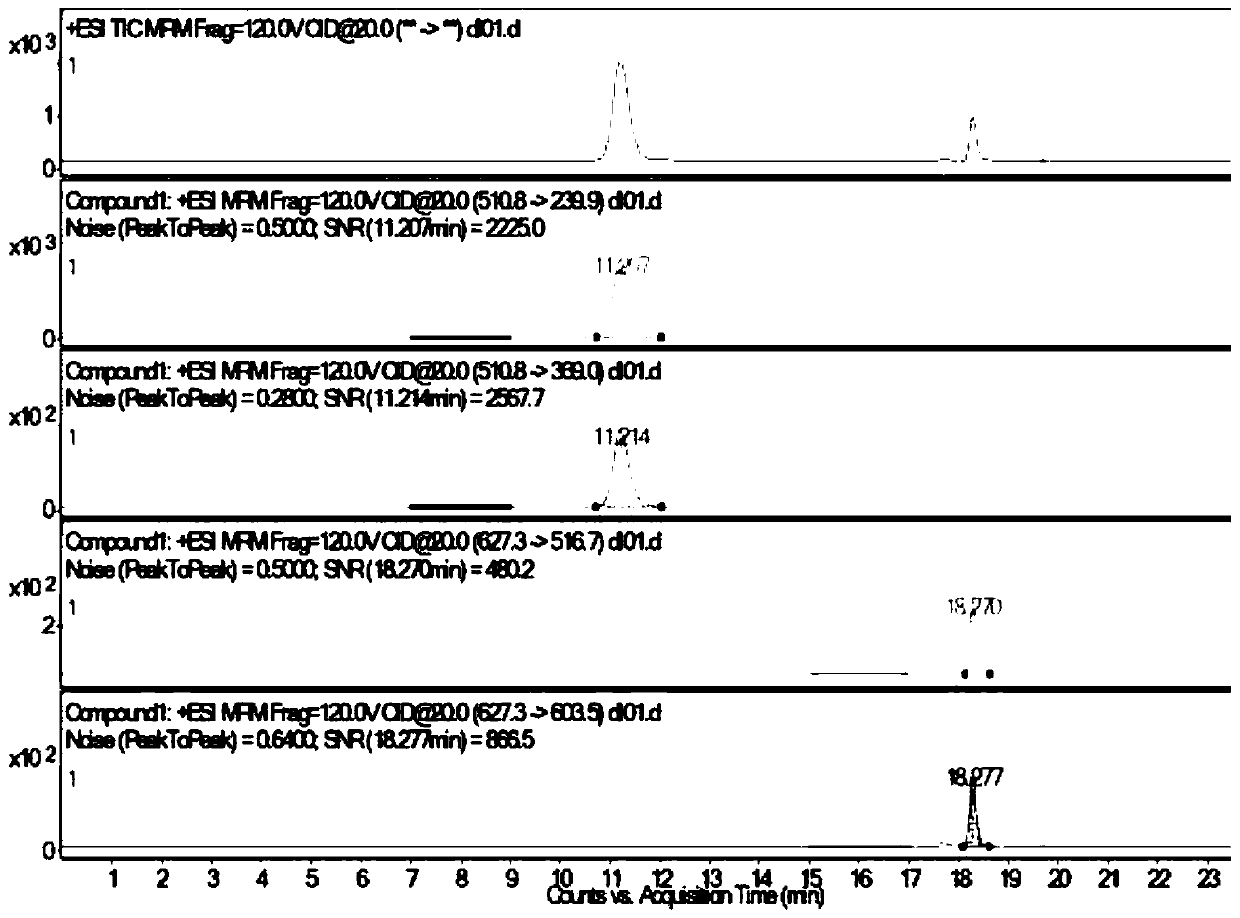
The invention discloses an identification method of periplaneta americana, which comprises the specific steps of: performing enzymolysis treatment on a sample to be detected to prepare a test sample solution; then performing liquid chromatography-mass spectrometry detection on the test sample solution to determine whether the sample to be detected is the periplaneta americana or not, wherein, an electro-spray positive ion mode is adopted to perform multiple reaction monitoring, and MRM (Multiple Reaction Monitoring) scanning is performed by characteristic peptide ion pairs. The identificationmethod of the invention has the beneficial effects that: based on the found characteristic peptide, multiple reaction monitoring of mass spectrum is adopted, and the periplaneta americana and a counterfeit product thereof can be distinguished through detection of the characteristic ion pairs; the identification method has strong characteristics and high sensitivity, can identify various counterfeit varieties of the periplaneta americana, and can be suitable for identifying related extracts and preparations which cannot be subjected to DNA identification and detection.
Owner:ZHEJIANG JINGXIN PHARMA +2
A Latent Fingerprint Visualization and Chemical Residue Detection Method Based on Upconversion Nanoparticles
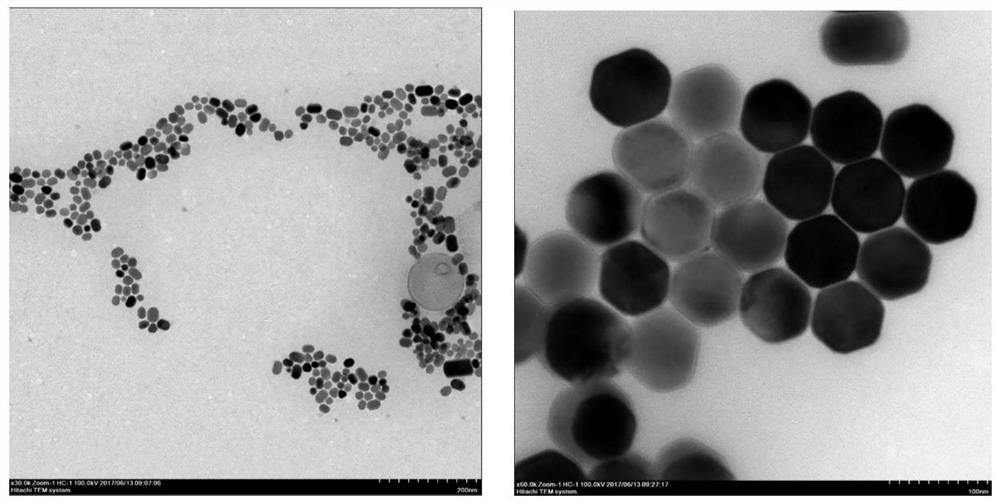
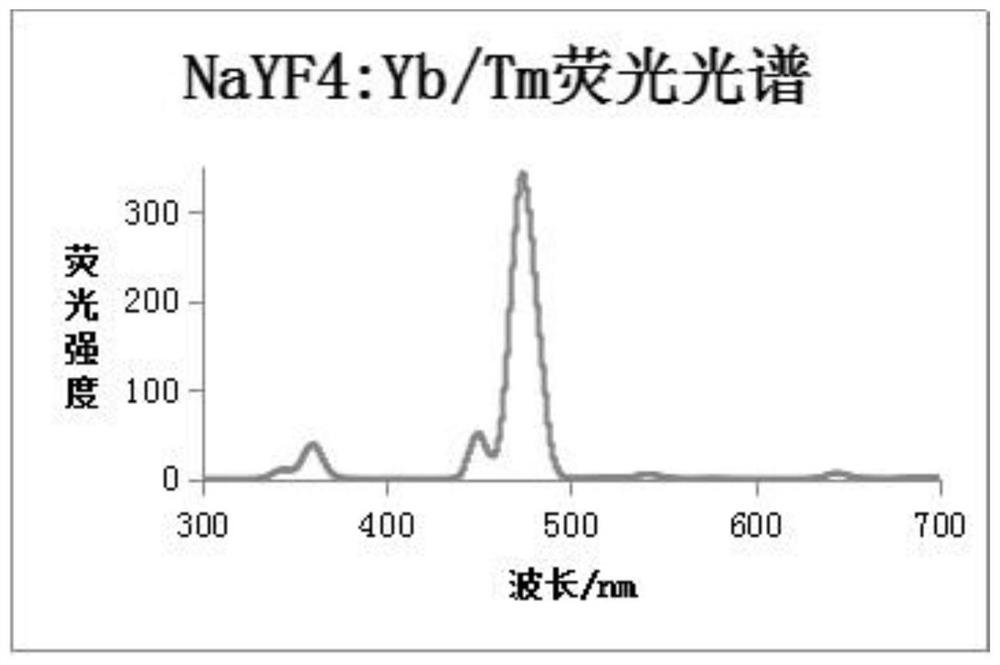
ActiveCN109001169BReduce distractionsLow background noiseFluorescence/phosphorescenceChemical compoundLatent fingerprint
The invention discloses a latent fingerprint development and chemical residue detection method based on upconversion nanoparticles. The method comprises the following steps: preparing the upconversionnanoparticles NaYF4:Yb<3+> / Tm<3+> with a hexagonal structure; preparing a latent fingerprint sample; forming a ligand compound; developing latent fingerprints. With the adoption of the upconversion nanoparticles, interference in the fingerprints is small, color development speed is higher, background interference is small, latent information of the latent fingerprints is not damaged, DNA identification of the latent fingerprints is not affected, toxic and side effects are avoided in synthesis and detection processes, nicotine development fluorescence change is obvious, sensitivity is high, detection equipment is portable, all devices used for the whole detection are portable and movable, and on-site detection is facilitated.
Owner:ZHEJIANG UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com