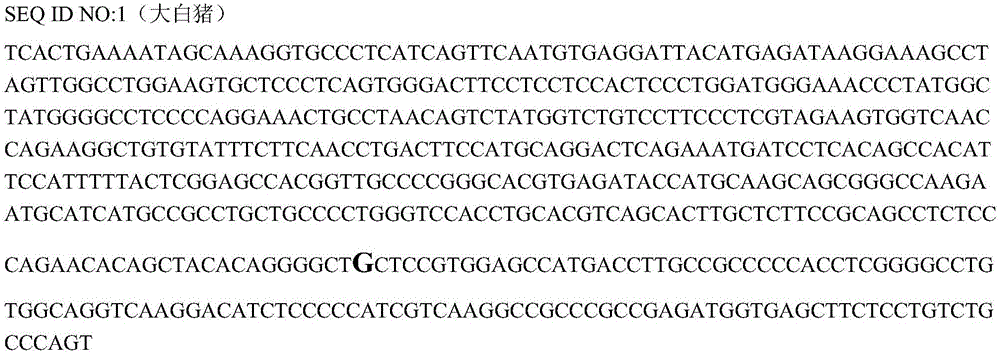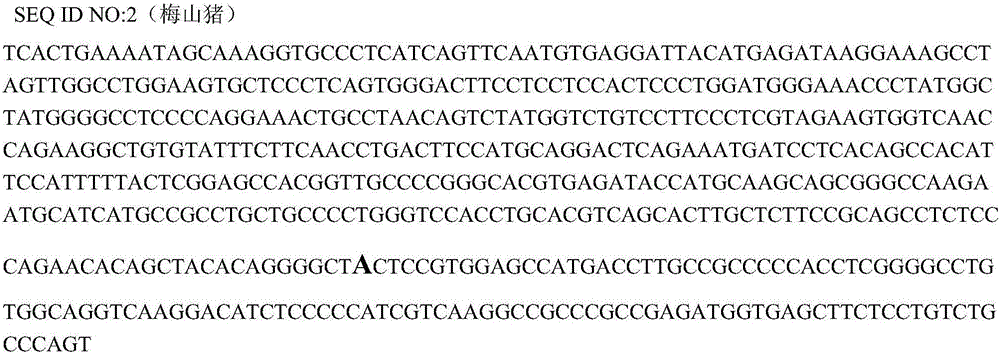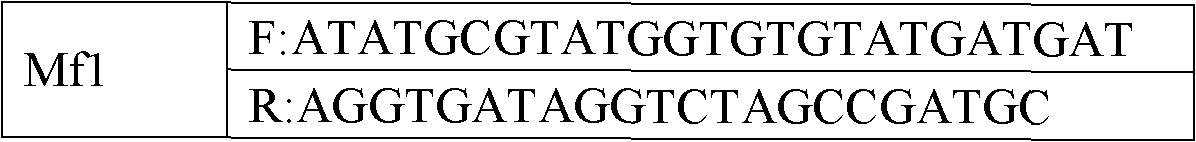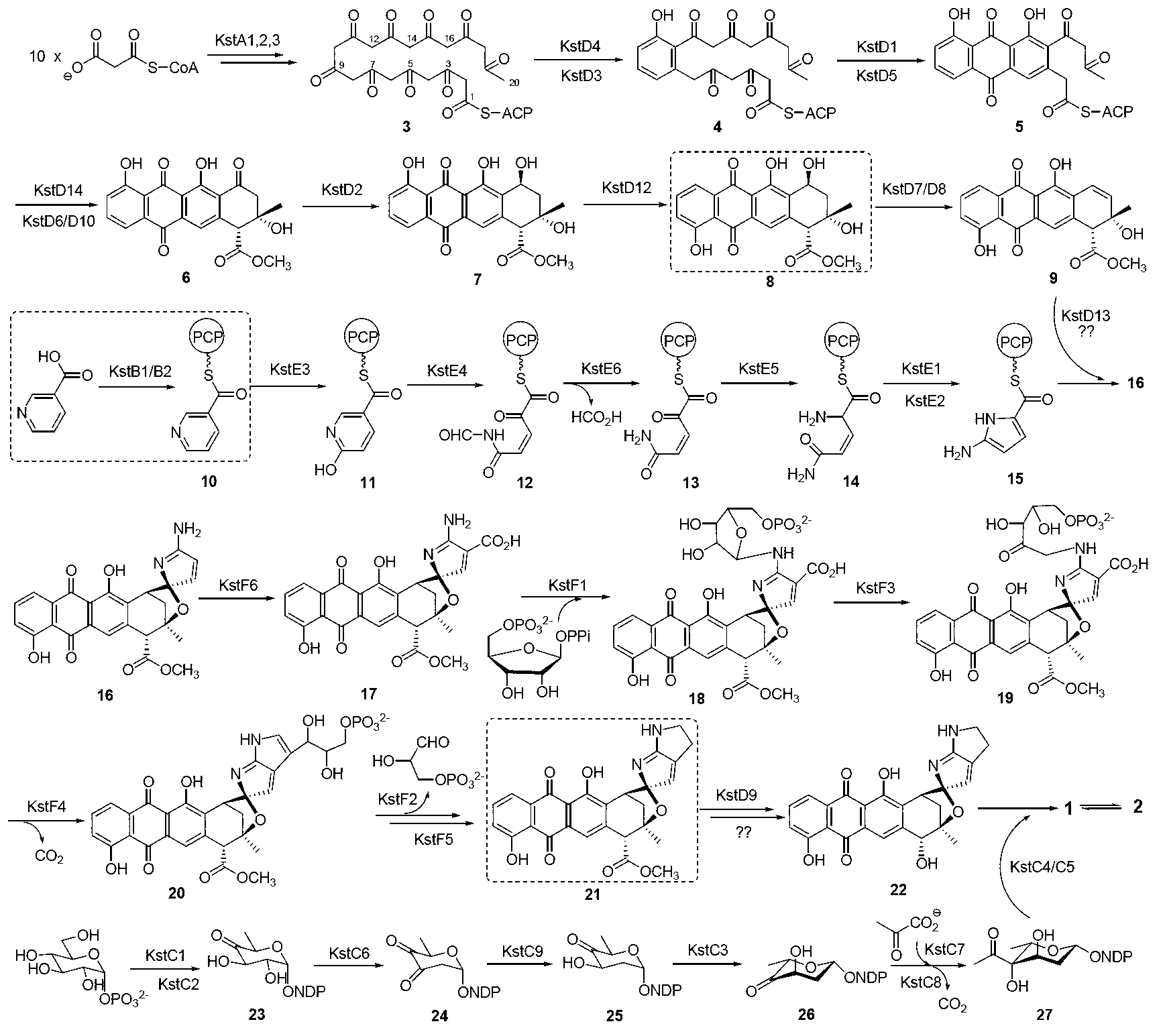Patents
Literature
199 results about "Cloning sequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Paired end sequencing
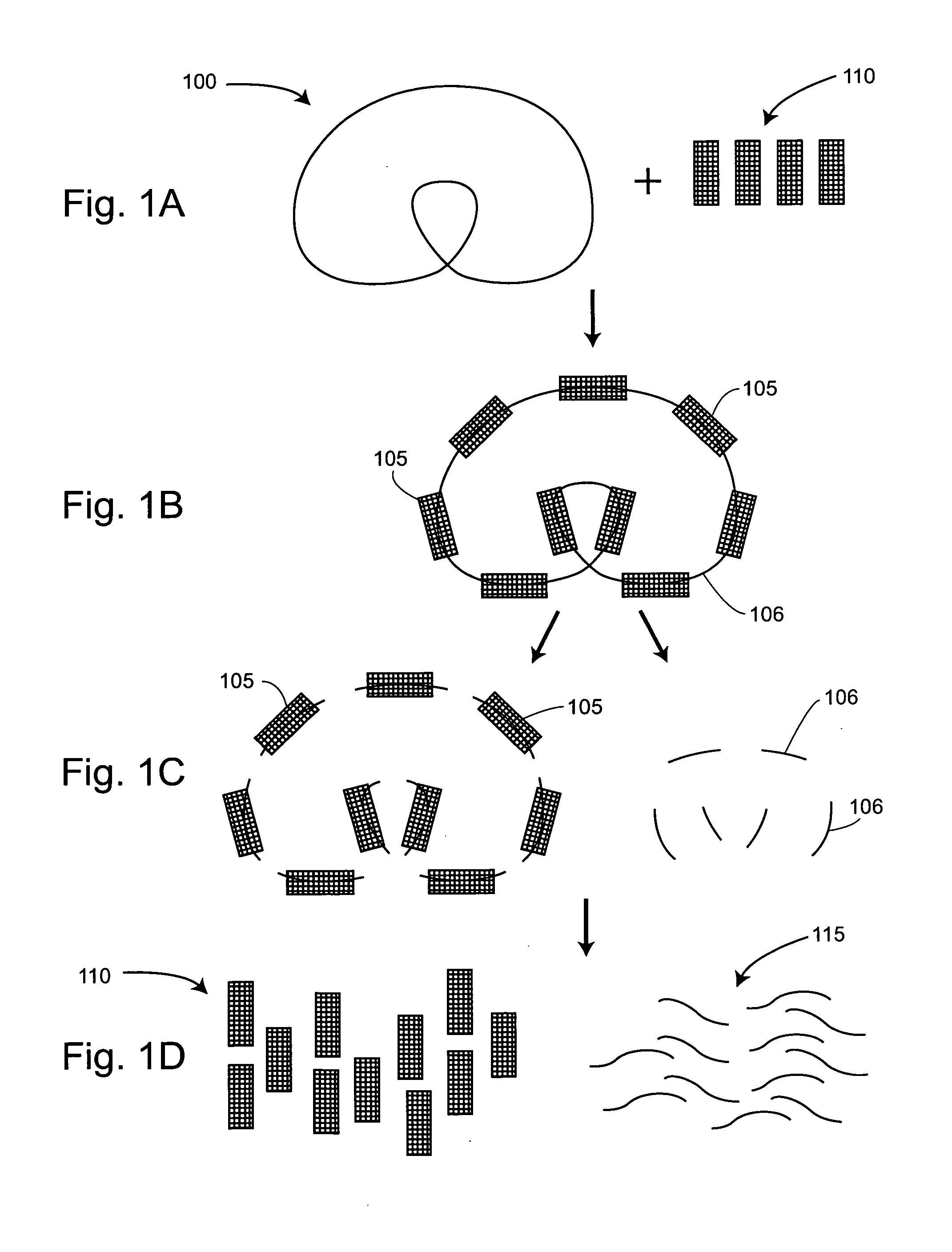
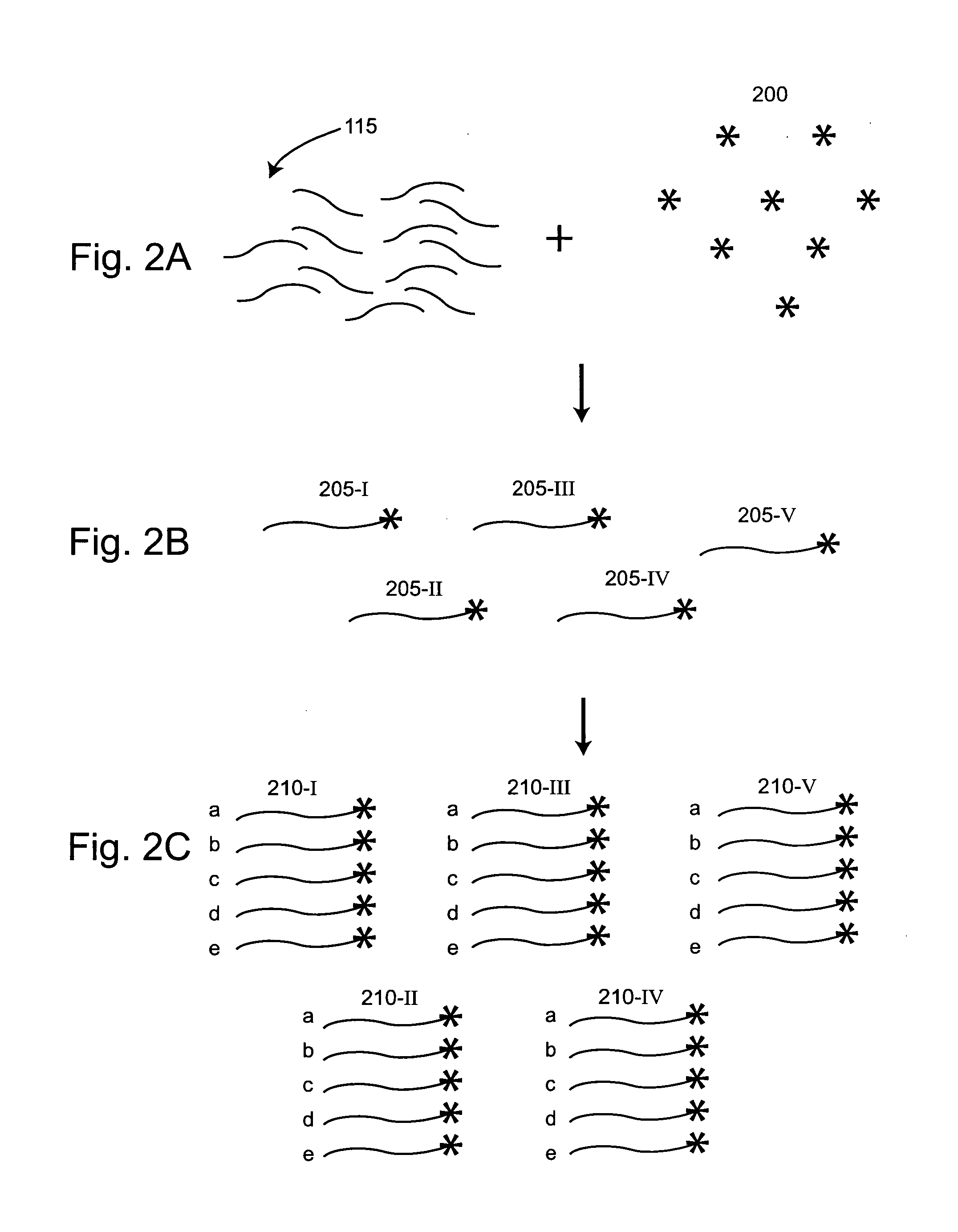
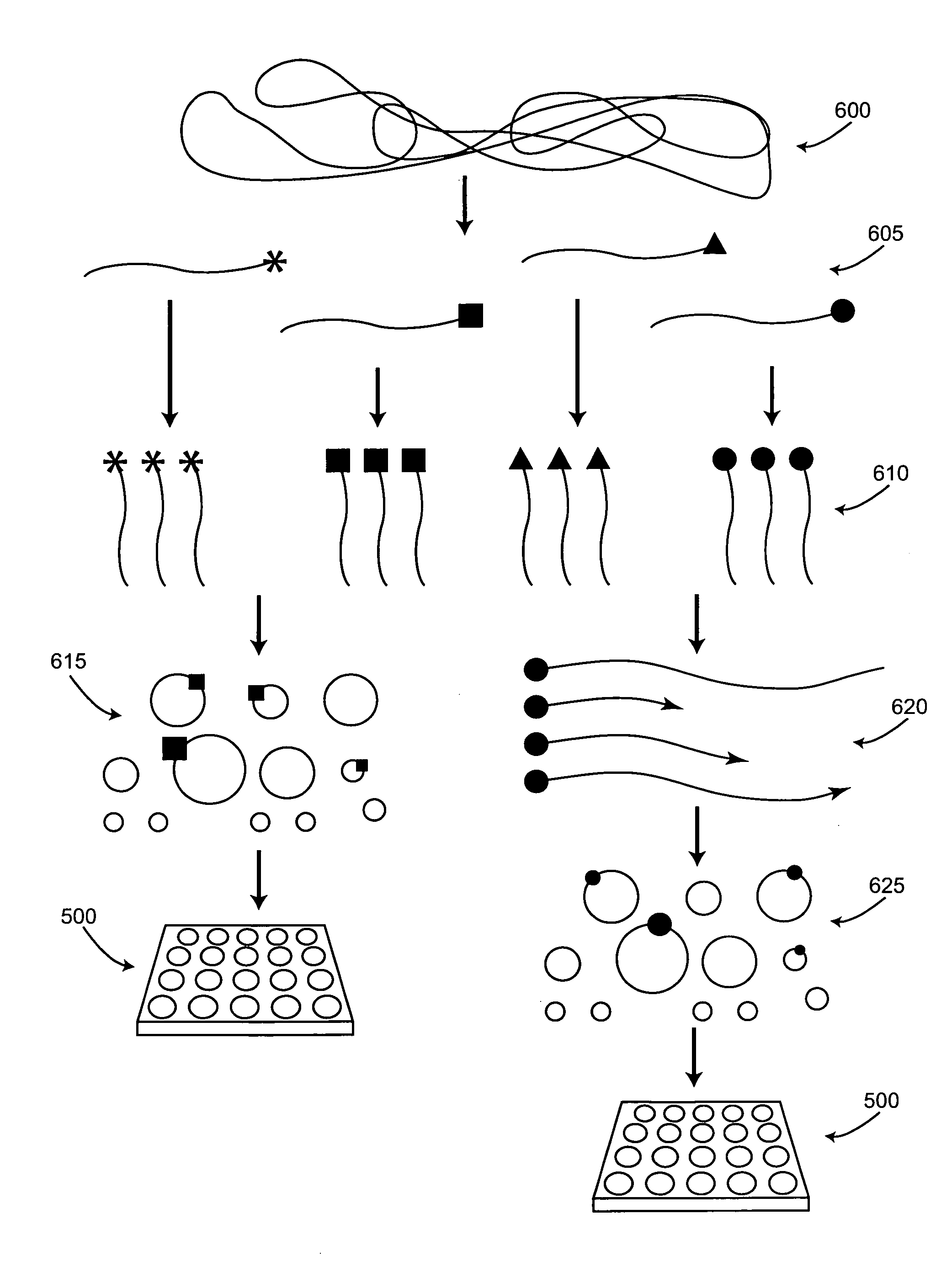
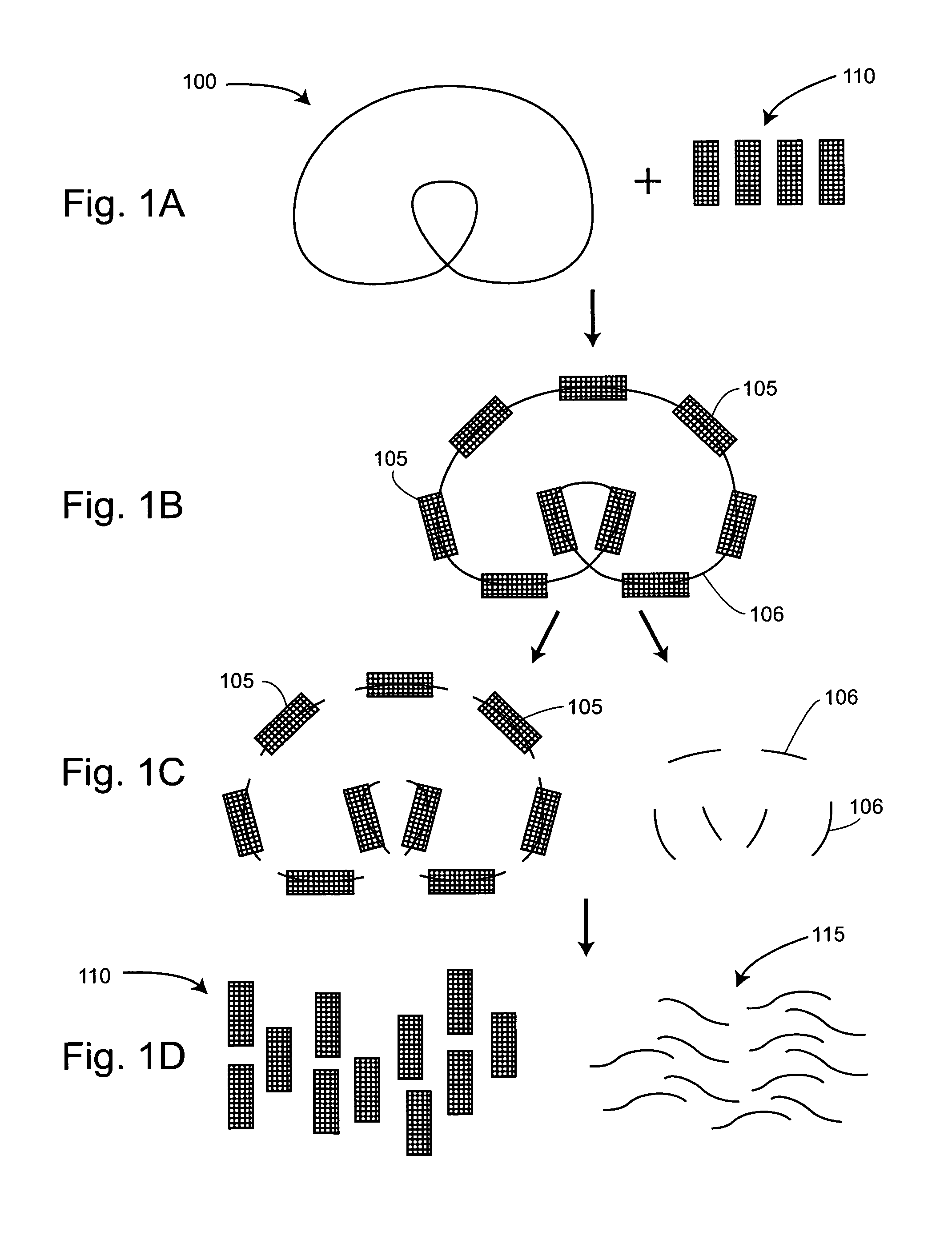
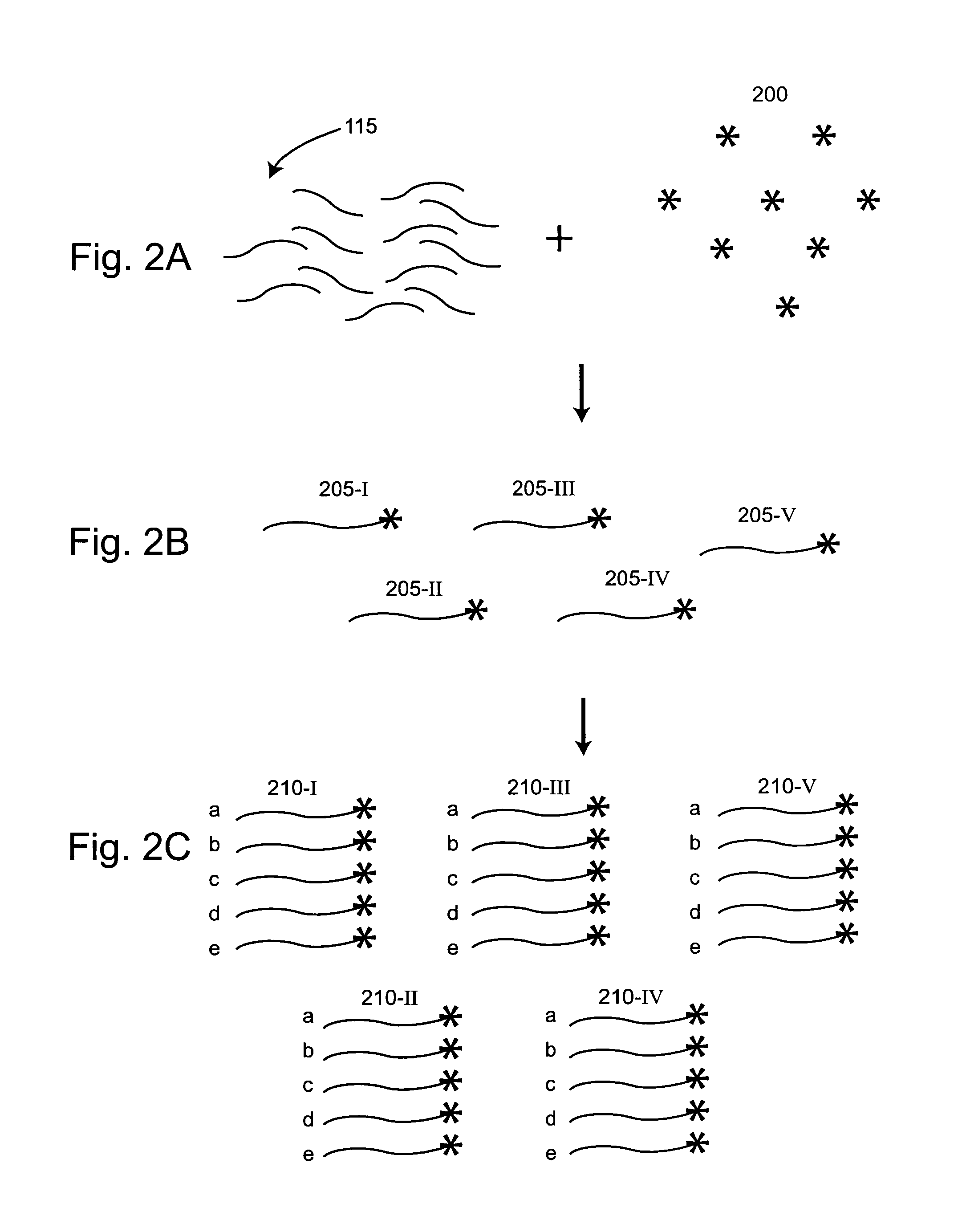
The present invention provides for a method of preparing a target nucleic acid fragments to produce a smaller nucleic acid which comprises the two ends of the target nucleic acid. Specifically, the invention provides cloning and DNA manipulation strategies to isolate the two ends of a large target nucleic acid into a single small DNA construct for rapid cloning, sequencing, or amplification.
Owner:454 LIFE SCIENCES CORP
Error-free amplification of DNA for clonal sequencing
ActiveUS20090105094A1Reduce probabilitySequencing errorMicrobiological testing/measurementFermentationCloning sequencingDNA
Methods of preparing nucleic acid templates and providing the nucleic acid templates to low copy number reaction volumes are provided. Related compositions of nucleic acid templates are also provided.
Owner:PACIFIC BIOSCIENCES
Error-free amplification of DNA for clonal sequencing
ActiveUS8003330B2Reduce probabilityMicrobiological testing/measurementFermentationCloning sequencingDNA
Provided are methods of producing low copy number circularized nucleic acid variants that can be distributed to reaction volumes. The methods include providing a template nucleic acid; producing a population of clonal nucleic acids from the template nucleic acid; generating a set of partially overlapping nucleic acid fragments from the population of clonal nucleic acids; circularizing the partially overlapping nucleic acid fragments to produce circularized nucleic acid variants; and aliquotting the circularized nucleic acid variants into reaction volumes. Related compositions of nucleic acid templates are also provided.
Owner:PACIFIC BIOSCIENCES
Environment DNA identification method for fish community structure researching
ActiveCN104531879AProtected speciesSave human and financial resourcesMicrobiological testing/measurementCommunity structureNucleotide sequencing
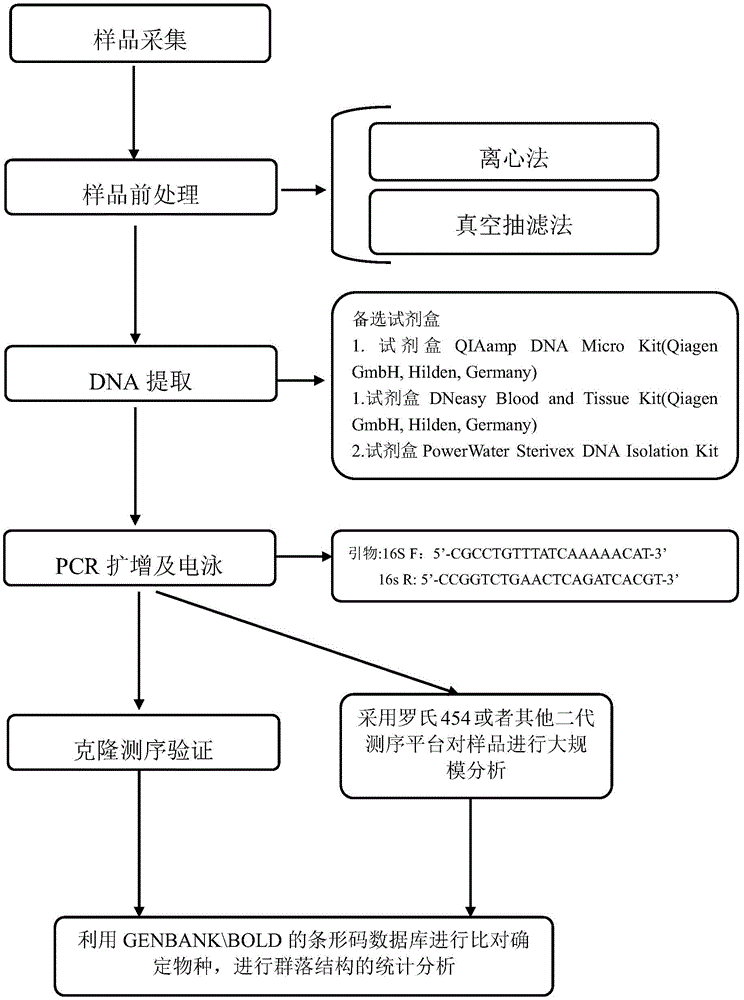
The invention relates to an environment DNA identification method for fish community structure researching. The method is characterized by comprising the steps that 1, water samples are collected according to the size of the water area where a fish community is located; 2, the water samples are processed, and total environment DNA of the processed water samples is extracted; 3, a 16s r DNA sequence with nucleotide sequences being SEQ ID NO:1 and SEQ ID NO:2 is adopted as a universal primer to carry out PCR amplification on the total environment DNA, and a PCR product is obtained; 4, gel electrophoresis is carried out on the PCR product, and gel with DNA is obtained; 5, gel cutting and clone sequencing are carried out on the gel with the DNA, and then blast comparison is carried out through GENBANK so as to determine whether the DNA is a fish sequence or not; 6, after it is determined that the DNA is the fish sequence, the next-generation sequencing technology is adopted for analyzing composition conditions of the PCR product on a large scale, and therefore the environment DNA fish composition and community structure are determined. The environment DNA identification method is quite easy, convenient and efficient and has the practical value.
Owner:SHANGHAI OCEAN UNIV
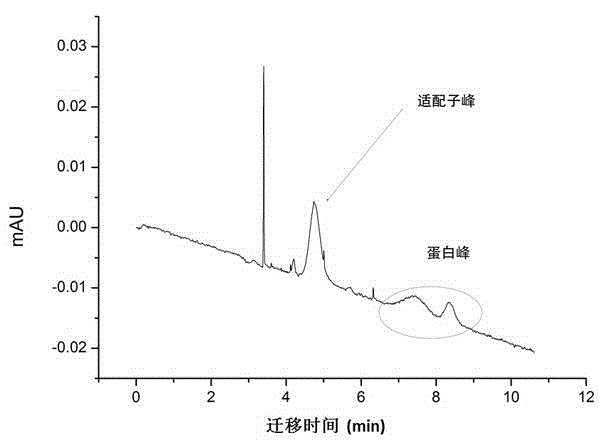
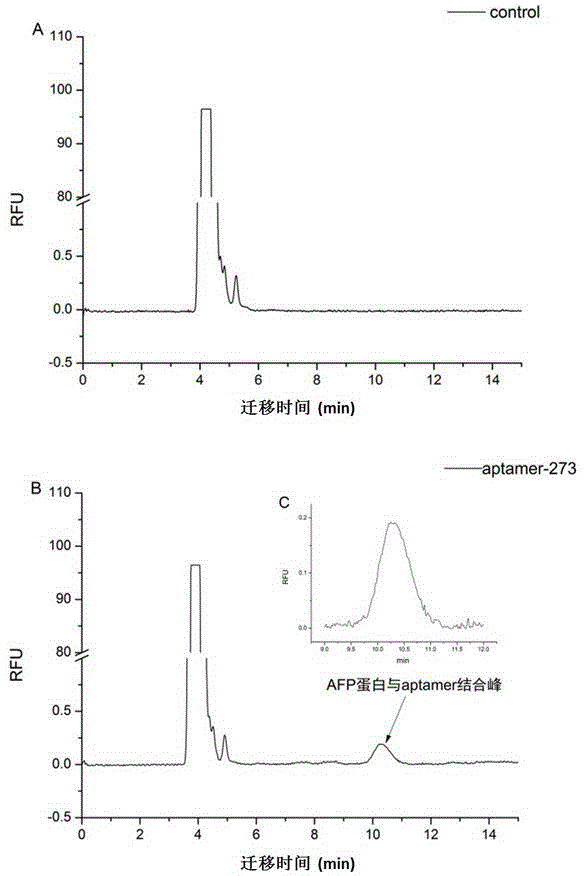
Method for screening aptamer specifically bound with alpha-fetoprotein
InactiveCN104131105AStrong interactionIncrease workloadMicrobiological testing/measurementDNA preparationAptamerFetuin b
Owner:ZHONGSHAN HOSPITAL FUDAN UNIV +1
Cucumber female deseription related molecule label and uses thereof
InactiveCN101113469AShorten breeding timeMicrobiological testing/measurementPlant genotype modificationAgricultural scienceRe engineering
The invention discloses a molecular marker related to female traits of cucumber. The molecular marker is two SNP divergence sites of EIN3 gene of cucumber happening in places of 153bp and 501bp, wherein, the EIN3 gene of pure female plant has molecular markers of G153bp and T501bp or has any one of molecular markers of G153bp and T501bp. The method for obtaining the molecular marker is that first DNA group of young leaf of cucumber is extracted and the primer is designed to do PCR amplification, then a cloning sequence test for PCR products and comparison and analysis of a sequence test result are done and finally the primer is redesigned in parents and F2 groups to be verified. The molecular marker can be taken as specific marker for selection of female cucumber and applied to assisting selection of cucumber sex for breeding through gene marker, which significantly shortens breeding time.
Owner:易克 +1
Three oligonucleotide sequences for identification and detection of vibrio alginolyticus as well as preparation method and application thereof
ActiveCN102329862AImprove stabilityQuick checkMicrobiological testing/measurementDNA preparationSingle strandOligonucleotide
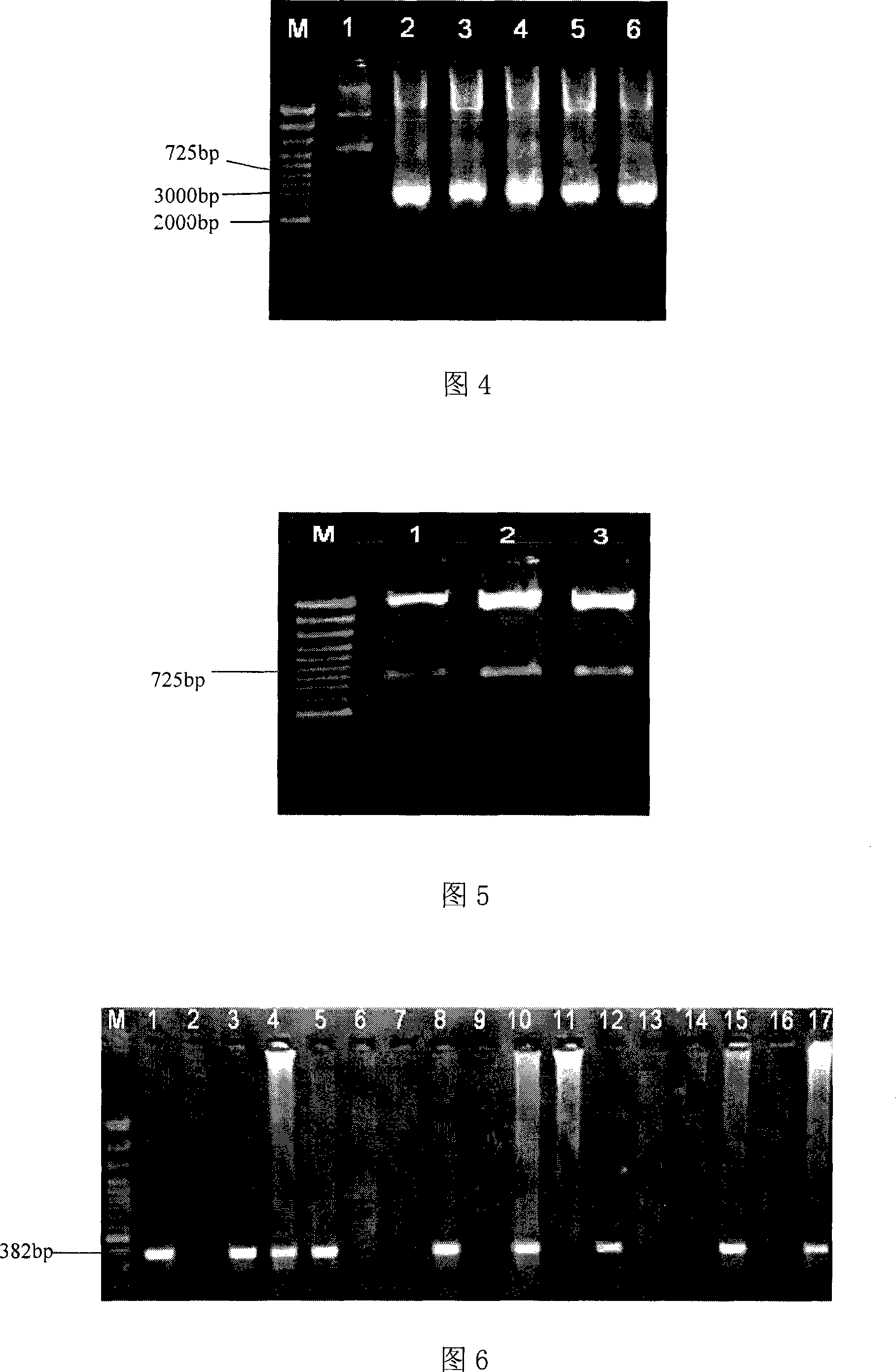
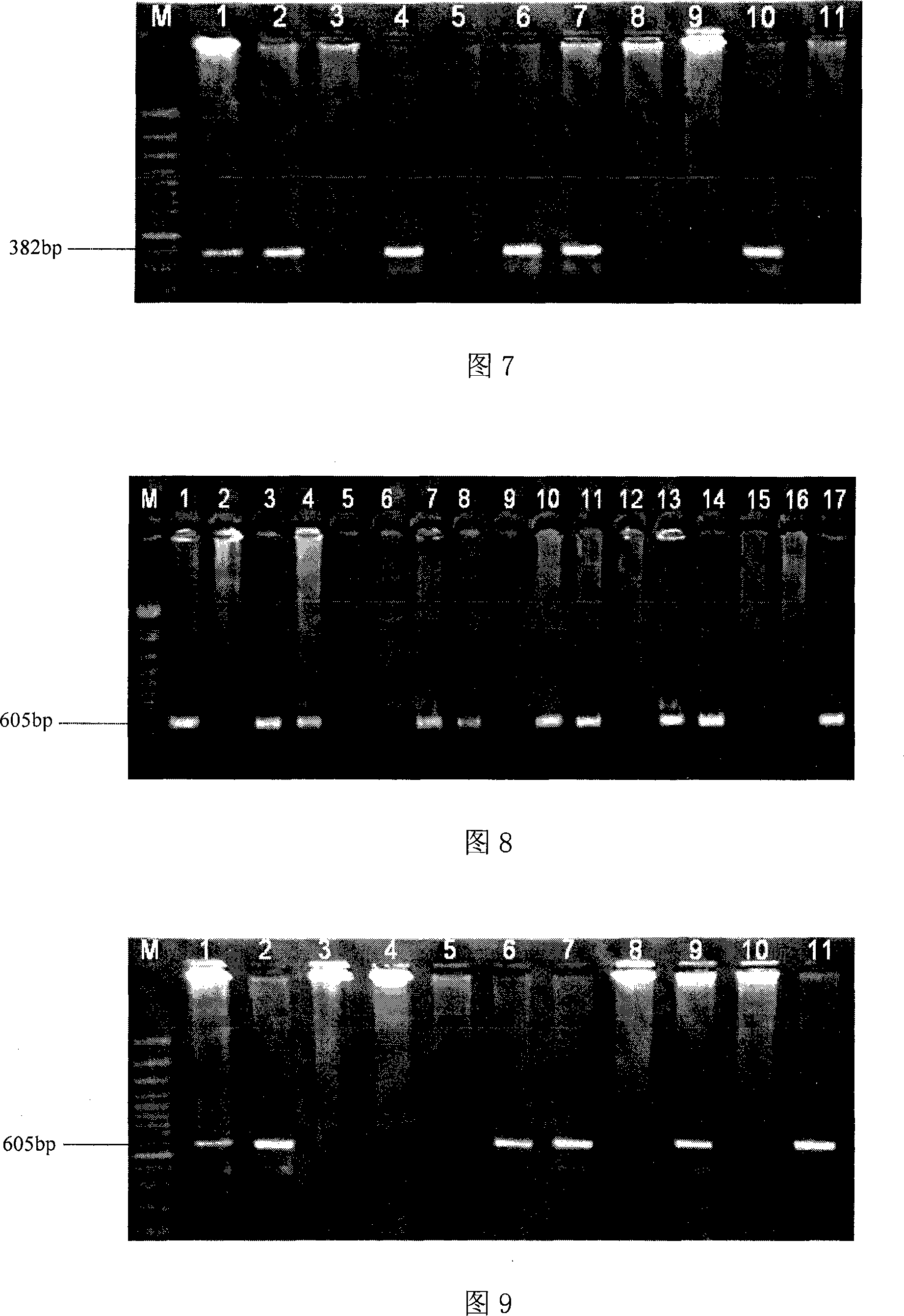
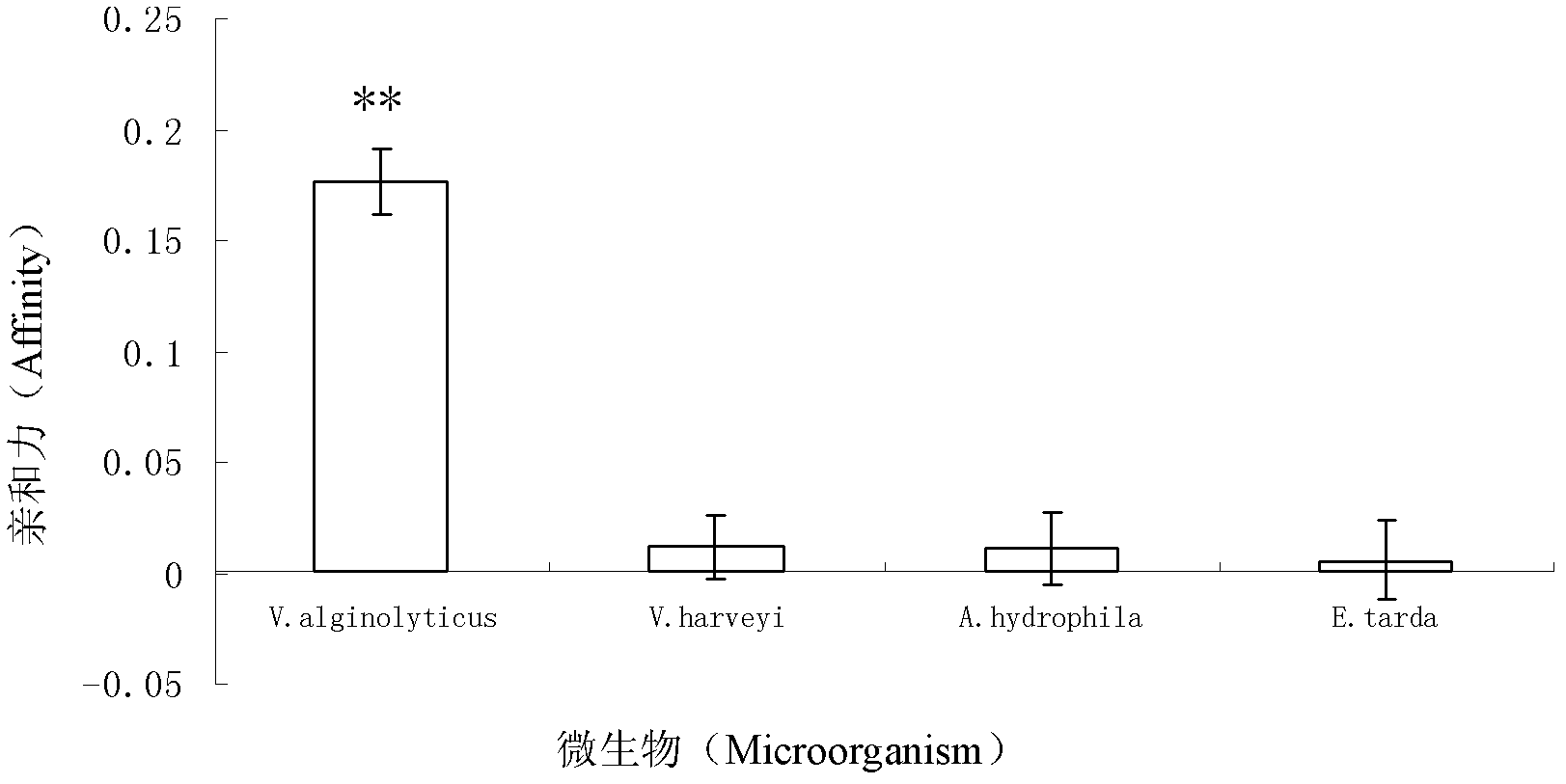
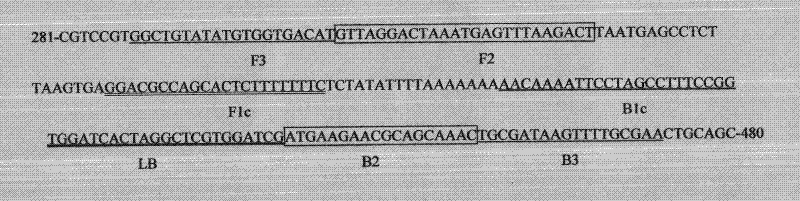
The invention discloses three oligonucleotide sequences for identification and detection of vibrio alginolyticus as well as a preparation method and an application thereof and relates to identification and detection of vibrio alginolyticus. The oligonucleotide sequences comprise SEQ ID No. 1-3 (sequence identity number 1-3). The method comprises the following steps: synthesizing an ssDNA (single-stranded deoxyribonucleic acid) oligonucleotide library (5'-TCA GTC GCT TCG CCG TCT CCT TC----N35----GCA CAA GAG GGA GAC CCC AGA GGG-3') for screening; mixing the oligonucleotide library with the vibrio alginolyticus and then performing SELEX (systematic evolution of ligands by exponential enrichment) screening; performing cloning and sequencing on an aptamer-enhanced library after completing the SELEX screening; performing interception in different lengths on high-copy ssDNA emerged in the sequencing result so as to get a series of new sequences, and further constructing complementary sequences of the new sequences; and performing affinity and specificity verification on the obtained new sequences and the complementary sequences thereof so as to get corresponding aptamers.
Owner:JIMEI UNIV
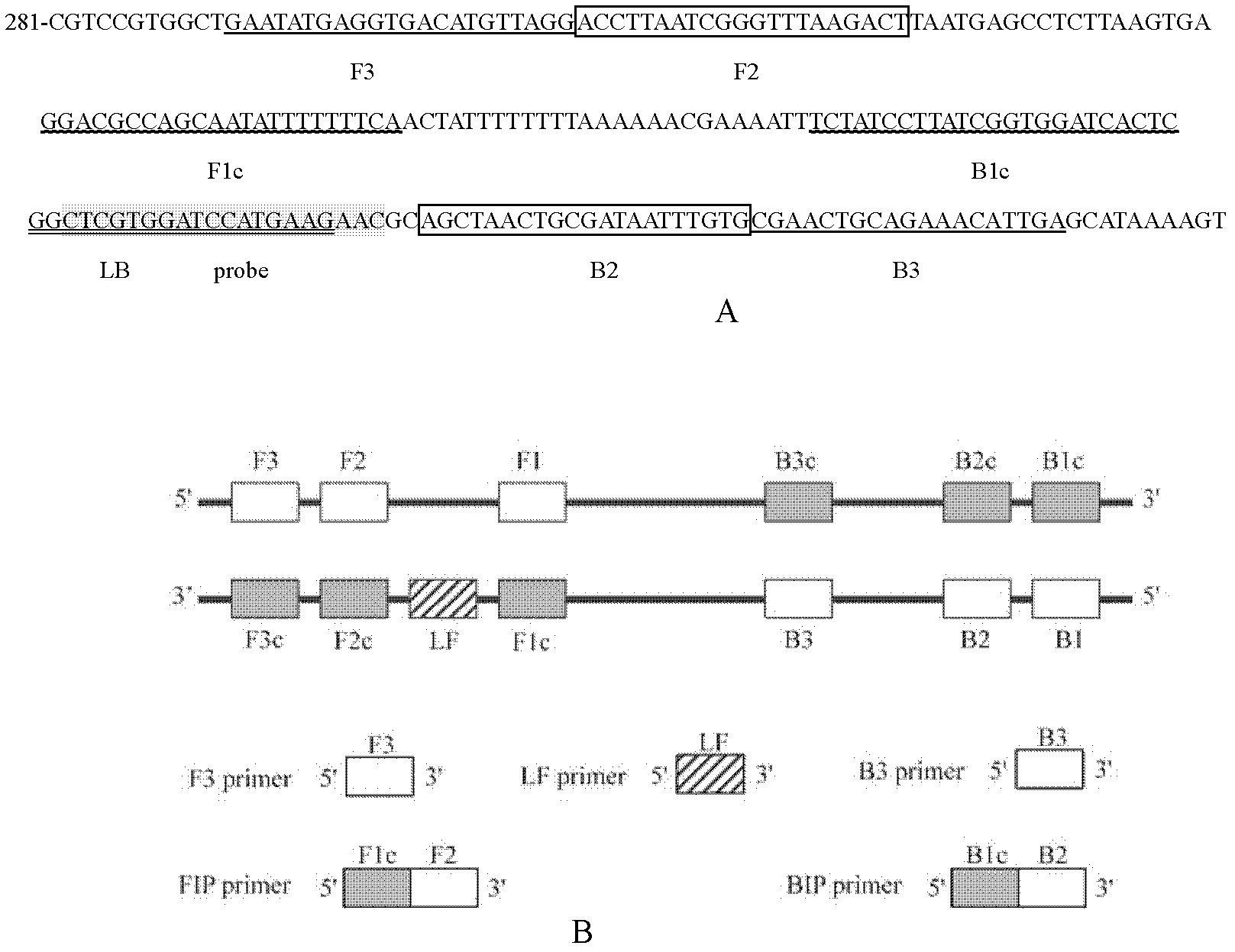
Enterolobium cyclocarpum knot nematode loop-mediated isothermal amplification (LAMP) rapid detection method and application
InactiveCN102174650AEasy to operateHigh sensitivityMicrobiological testing/measurementLoop-mediated isothermal amplificationDNA extraction
The invention relates to an enterolobium cyclocarpum knot nematode loop-mediated isothermal amplification (LAMP) rapid detection method and applications. The method comprises the following steps: designing 5 LAMP primers: MEF3, MEB3, MEFIP, MEBIP and MELB in a sequence conversed domain according to an enterolobium cyclocarpum knot nematode ITS sequence of the clone sequencing; configuring a LAMP reaction system; detecting isothermal amplification and amplification product color developing of the enterolobium cyclocarpum knot nematode through extracting the DNA of the enterolobium cyclocarpum knot nematode, thus rapidly detecting the enterolobium cyclocarpum knot nematode. The detection method provided by the invention has strong specific, high sensitivity, low cost and simple operation, and has high application value in the aspects of spot rapid detection and the early-stage diagnosis of the enterolobium cyclocarpum knot nematode.
Owner:INST OF PLANT PROTECTION CHINESE ACAD OF AGRI SCI
Genetic marker related to porcine semen quality traits and application
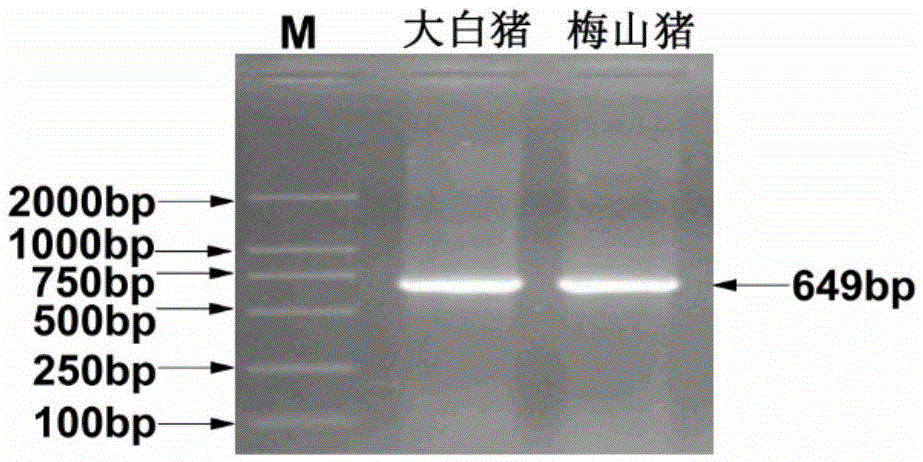
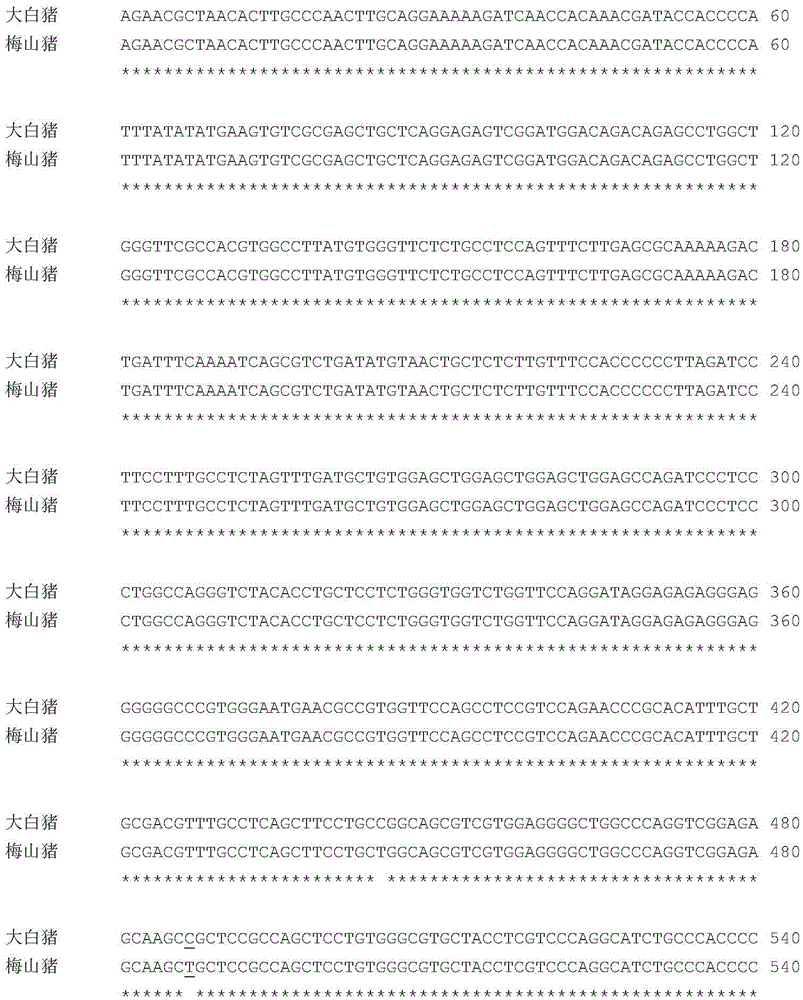
The present invention belongs to the technical field of screening and application of pig genetic markers, and in particular relates to a genetic marker related to porcine semen quality traits and application, the genetic marker is a gene obtained by cloning of a pig RNF4 gene 3 'untranslated region, the single nucleotide polymorphism (SNP) can be used as a genetic marker of the porcine semen quality traits, a nucleotide sequence of the genetic marker is as shown in SEQ ID NO: 3 or SEQ ID NO: 4, the sequence SEQ ID NO: 3 or SEQ ID NO: 4 has an allelic gene mutation at 487bp, and AluI-RFLP polymorphism is caused by the allelic gene mutation. Screening step of the method are as follows: extraction of genomic DNA from pig blood, primer design, PCR amplification, PCR product cloning, sequencing, sequence comparing analysis, single nucleotide polymorphism detection, marking and semen quality trait correlation analysis. The genetic marker can be used in pig marker assisted selection.
Owner:HUAZHONG AGRI UNIV
Rapid molecule detecting method for microflora composition in waste water biological treatment reactor
InactiveCN101475987AAccurate analysisMonitoring timeMicrobiological testing/measurementBiological water/sewage treatmentGenomic DNAGel electrophoresis
A rapid molecular detection method of microbial community composition in a wastewater biological treatment reactor includes: collecting the microorganism samples in the wastewater biological treatment reactor for pretreatment, and using the phenol-chloroform method for extracting total genomic DNA; using 16S rDNAV3 area primers to perform PCR amplification to the sample DNA; using denaturing gradient gel electrophoresis to analyzing the PCR amplification products; using software to analyze the obtained DGGE patterns, and identifying the represented microorganism based on the gel cutting sequencing feature DGGE bands; designing and constructing denaturing gradient gel electrophoresis database as the contradistinguish standard of rapid molecular detection based on the microorganism kinds represented by bands, wherein, the denaturing gradient gel electrophoresis integrated database is designed and constructed by gel cutting cloning sequencing the bands after the PCR products is processed after the DGGE separation and then defining the represented microorganism kinds after consanguinity comparison in the Genbank.
Owner:NANJING UNIV
Broad-spectrum nucleic acid aptamer capable of specifically identifying lipopolysaccharides and directed screening method of broad-spectrum nucleic acid aptamer
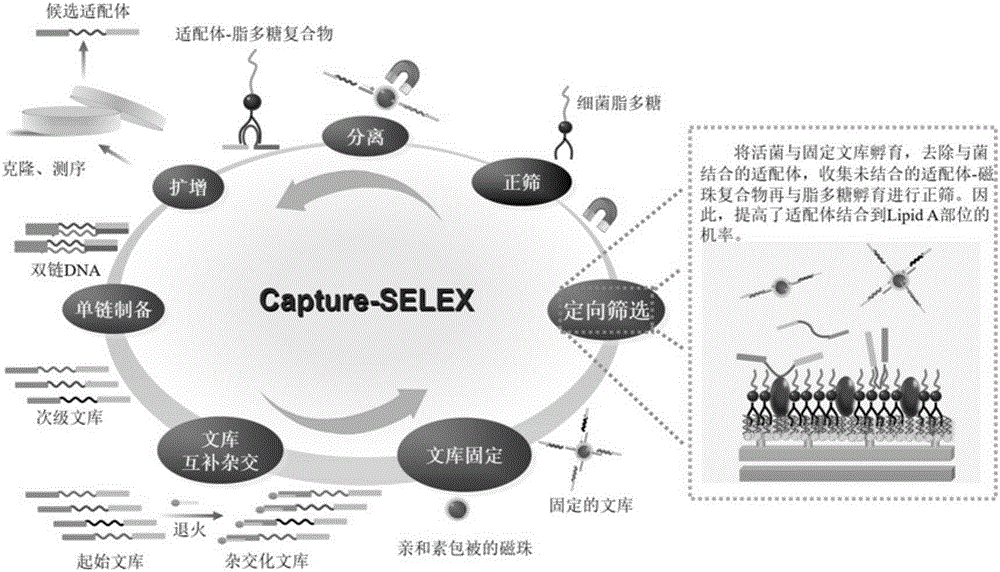
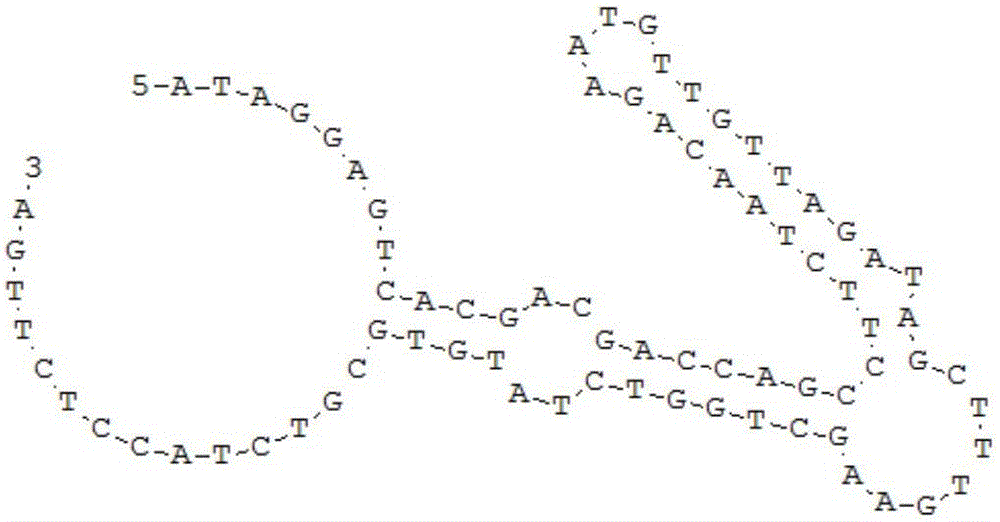
The invention provides a broad-spectrum nucleic acid aptamer capable of identifying lipopolysaccharides of different Gram-negative bacterium sources. The broad-spectrum nucleic acid aptamer is obtained by directed screening on the basis of the capture-SELEX technology of label-free target molecules and immobilized nucleic acid libraries. A directed screening method of the broad-spectrum nucleic acid aptamer includes: using mouse salmonella typhi lipopolysaccharides as the unique target, using a biotin-avidin effect to fix random oligonucleotide libraries to Fe3O4 magnetic nano particles wrapped by avidin through biotinylated short complementary chains, performing incubation, separation, amplification, digestion single chain preparation, cloning sequencing and the like, adding complete active bacteria of salmonella and escherichia coli to serve as directed molecules to perform directed screening when the libraries are enriched to a certain degree, and one broad-spectrum nucleic acid aptamer capable of identifying four kinds of lipopolysaccharides is obtained through 15 rounds of repeated screening. The nucleic acid aptamer is applicable to the analysis and detection, separation and enriching, toxicity neutralizing and the like of the lipopolysaccharides in aspects of drinking water, food or clinical medicine and the like.
Owner:JIANGNAN UNIV
Recombinant porcine epidemic diarrhea virus (PEDV) lactococcus lactis and its construction method and use
InactiveCN103074291AGood immune protectionPromote absorptionBacteriaMicroorganism based processesProtective antigenWestern blot
Owner:DALIAN UNIVERSITY
Loop mediated isothermal amplification (LAMP) detection method for meloidogyne hapla and application of method
The invention relates to a loop mediated isothermal amplification (LAMP) detection method for meloidogyne hapla and application of the method and belongs to the field of biological technology. 5 LAMP primers (namely, MHF3, MHB3, MHFIP, MHBIP and MHLB) are designed in the sequence conservative region according to the cloning sequencing meloidogyne hapla ITS sequence; the 5' end of the probe MH-FITC-Probe is marked by FITC; and the 5'end of the MHFIP primer is marked by the biotin. The DNA of the meloidogyne hapla is extracted, isothermal amplification is performed by the mixture of the specific primers, reaction buffer liquor and reaction enzyme, and the amplification product is subjected to color developing detection, so that the meloidogyne hapla can be detected quickly. The detection method is high in specificity, high in sensitivity, low in cost, and simple and convenient to operate, and has a high application value in the aspects of on-site quick detection on the meloidogyne haplaand early diagnosis on the meloidogyne hapla disease.
Owner:INST OF PLANT PROTECTION CHINESE ACAD OF AGRI SCI
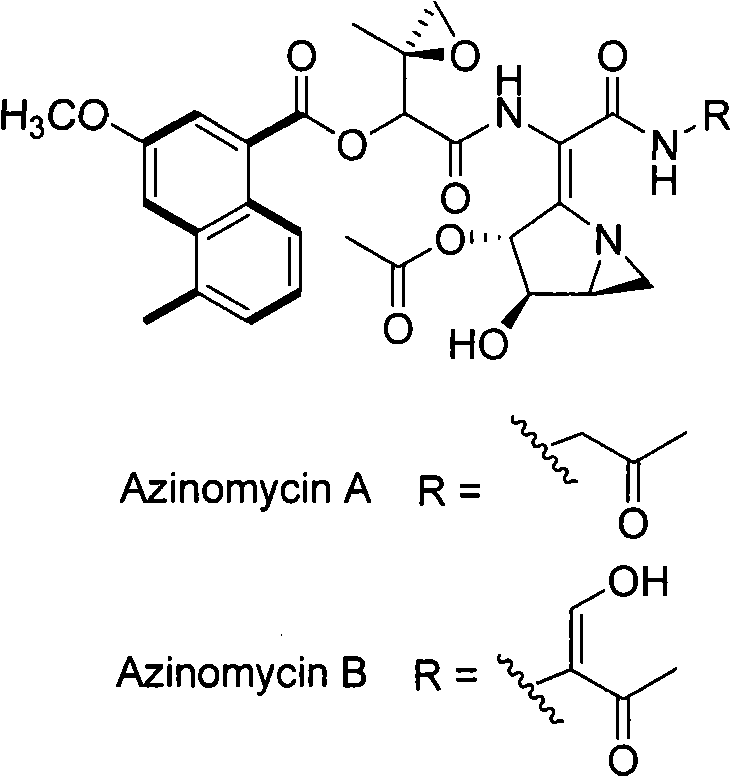
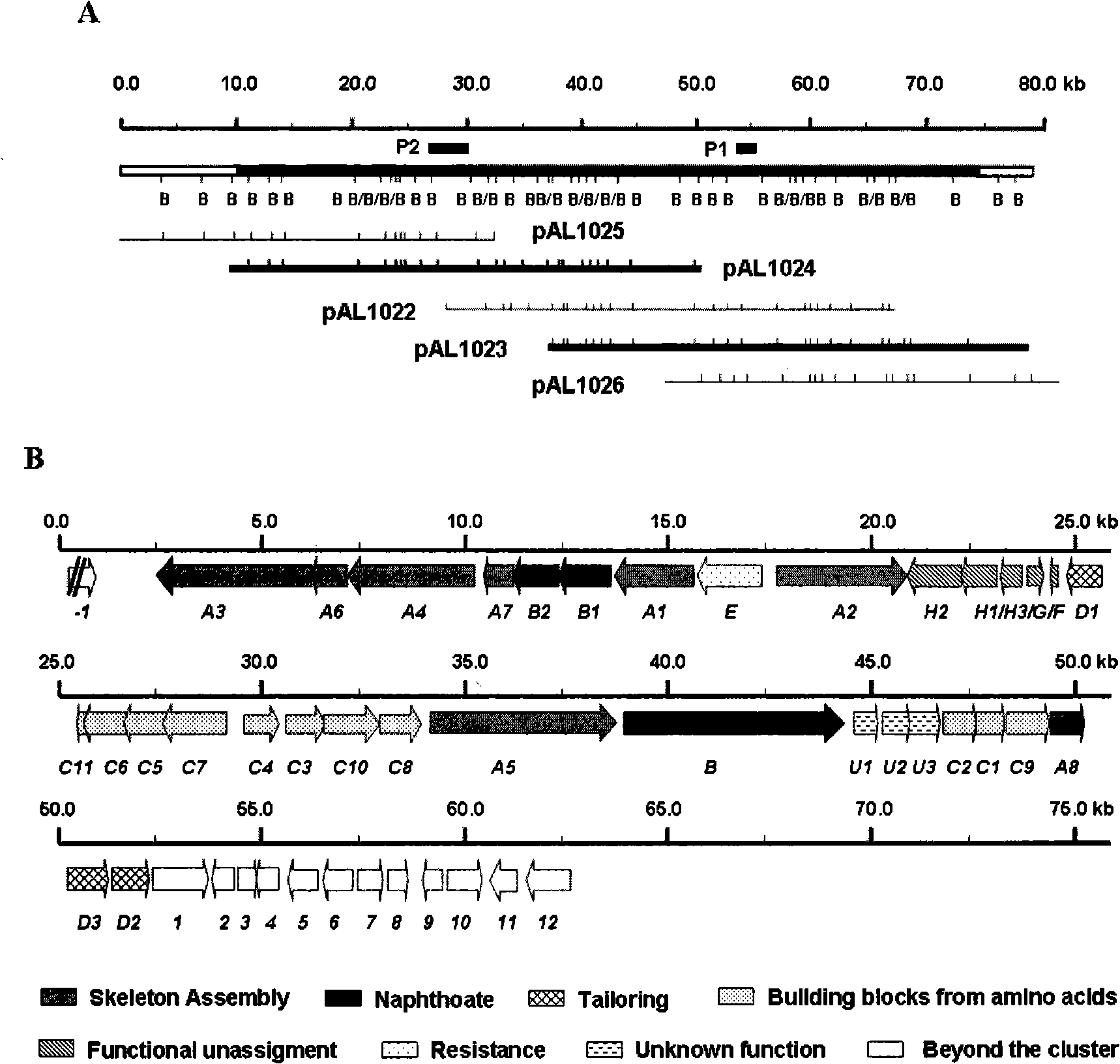
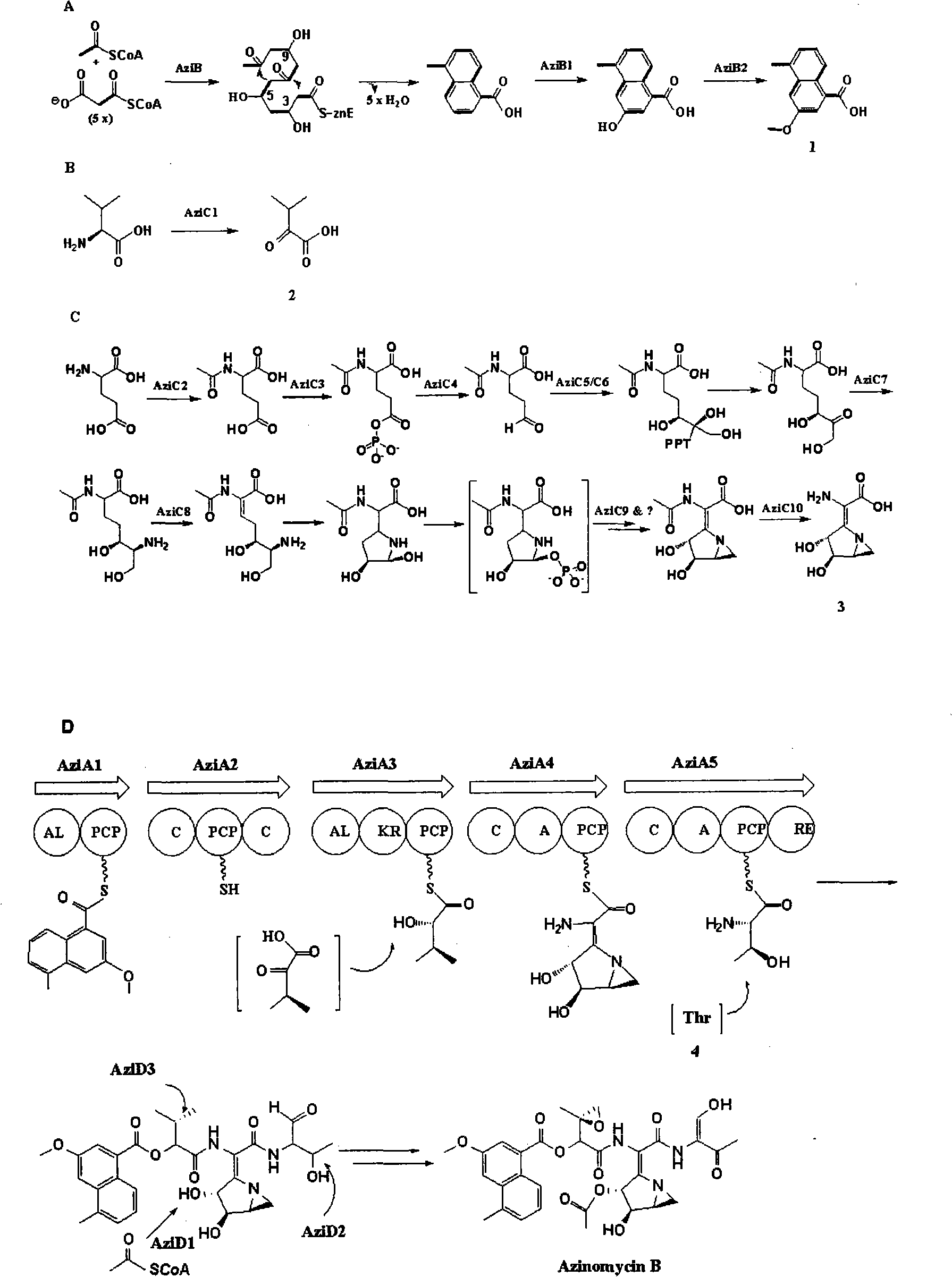
Biological synthesis gene cluster for Azintamide
InactiveCN101275141AUnderstanding Biosynthetic MechanismsFermentationPlant genotype modificationHeterologousEnzyme Gene
The present invention provides cloning sequencing, analyzing, function research of a biosynthesis gene cluster of an antibiotic-Azinomycin B having antitumor activity produced by streptomyces, and its application. The whole gene cluster includes 34 genes: one repeatedly using I type polyketide synthase gene; two naphthalene ring modification enzyme genes; 8 non-ribosomal polypeptide skeleton synthesis and modification enzyme genes; 11 non-natural amino acid structure unit synthase genes; 1 resistance gene; 3 post modification enzyme genes and 8 genes which functions are not determined. The genetic operation of the biosynthesis gene breaks the synthesis of Azinomycin B; the precursor compound is produced by the heterologous expression of synthesis gene and modification gene of naphthalene ring. The gene of the invention and the protein can be used for searching and finding compound or gene, protein applied in medical, industry or agriculture.
Owner:SHANGHAI INST OF ORGANIC CHEM CHINESE ACAD OF SCI
Method for analyzing Bacillus community structure in white spirit fermentation system
The invention discloses a method for analyzing the Bacillus community structure in a white spirit fermentation system, belonging to the technical field of microbial ecology. According to the invention, the diversity of a Bacillus community structure in the white spirit fermentation system is semi-quantitatively analyzed by a nested PCR-DGGE (polymerase chain reaction-denaturing gel gradient electrophoresis) method. The method comprises the following steps of: extracting the total genome DNA (deoxyribonucleic acid) in a yeast or fermented grains sample by a bead milling method; performing two steps of amplification of the Bacillus specific segment through the nested PCR; performing DGGE electrophoretic analysis; determining the microorganism species corresponding to the DNA stripe by use of a standard strain; performing glue cutting of the unknown stripe and recycling and then performing PCR again; connecting the T vector and cloning; performing DGGE electrophoresis again; performing positive cloning sequencing and performing blast comparison in the GenBank databae to obtain the microorganism information; digitally processing DGGE map by use of quantityone software; and calculating the proportion of different Bacillus strains in the total Bacillus flora according to the brightness of the stripe. The method disclosed by the invention has the advantages of simplicity, strong specificity and good repeatability, and lays foundation for further studying the importance of Bacillus in white spirit production.
Owner:JIANGNAN UNIV
Genetic marker associated with pig meat quality characters and carcass characters
InactiveCN105755131AMicrobiological testing/measurementDNA/RNA fragmentationMarker-assisted selectionIntramuscular fat
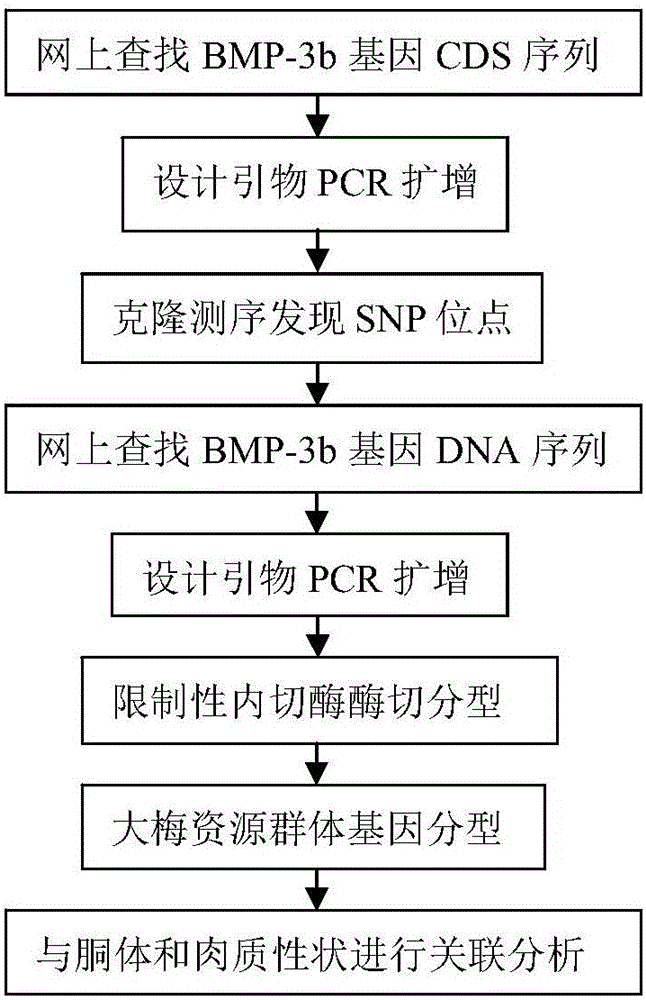
The invention belongs to the technical field of pig genetic marker preparation and particularly relates to a genetic marker associated with pig meat quality and carcass characters, wherein the marker is obtained by screening the pig BMP-3b gene, and the sequence is shown by SEQ ID NO:1 and figure 4; and A / G base mutation happens at the 425th-site base of the sequence and causes AlwNI-RFLP polymorphism. The screening method comprises the following steps: extracting genome DNA from pig blood; designing a primer; performing PCR amplification; cloning the PCR product and sequencing; comparing and analyzing the sequence; performing SNP detection; and analyzing the correlation between the marker and the meat quality characters. The invention discloses an SNP typing detection technology for the CDS area of the pig BMP-3b gene, and significant correlation is discovered between the A / G mutation site and the meat quality characters such as marbling, color value and intramuscular fat content and between the A / G mutation site and the carcass characters such as average back-fat thickness, eye muscle height and leaf fat. The invention provides a new marker resource for pig marker assisted selection.
Owner:HUAZHONG AGRI UNIV
Preparation method and use of simple sequence repeat (SSR) marker for screening miscanthus by magnetic bead enrichment process
InactiveCN102080130AEfficient enrichmentCo-dominance is goodMicrobiological testing/measurementMagnetic beadGenetic diversity
The invention discloses a preparation method and use of a simple sequence repeat (SSR) marker for screening miscanthus by a magnetic bead enrichment process, which comprises: 1) extracting genomic DNA of miscanthus floridulus which belongs to miscanthus; 2) incising the extracted total DNA by using restriction incision enzyme to obtain a genomic DNA; 3) intercrossing a probe and a target fragment; 4) enriching the hybrid DNA molecules having an SSR sequence by using magnetic beads; 5) amplifying and purifying the DNA fragment having the SSR sequence; 6) cloning and sequencing the DNA fragment; and 7) designing an SSR primer according to the flanking sequence of SSR, and designing a specific primer by using primer design software for amplifying the satellite fragment at the locus. The method is easy and convenient for operation; meanwhile, the SSR marker has the characteristics of high codominance, high polymorphism, multiple allele property and the like and can be used for analyzing the genetic diversity and genetic relationship of miscanthus and used in construction of genetic map of miscanthus, calibration of a target gene and drawing of a fingerprint.
Owner:湖北光芒能源植物有限公司
Process of cloning new resistance gene of wild rice
InactiveCN101045928AQuick searchOvercome the cumbersome processSugar derivativesMicrobiological testing/measurementOryzaAgricultural science
The present invention discloses process of cloning new resistance gene of wild rice. The process includes the first constructing library of convertible genomes of donor wild rice, the subsequent designing primer according to the conservation sequences of the cloned rice blast resisting gene and bacterial leaf spot resisting gene to obtain amplified product as the probe, performing bead enrichment on the library with the marked probe to establish candidate resistance gene enriching library, and the final spot hybridization analysis and PCR identification on the cloning of the enriching library, the cloning sequencing analysis and function verification of the amplified product. The present invention integrates candidate resistance gene cloning technology and bead enriching library constructing technology, and is suitable for cloning of wild rice resistance gene and other beneficial plant gene.
Owner:HUNAN WEST CITY HYBRID RICE GENE TECH +1
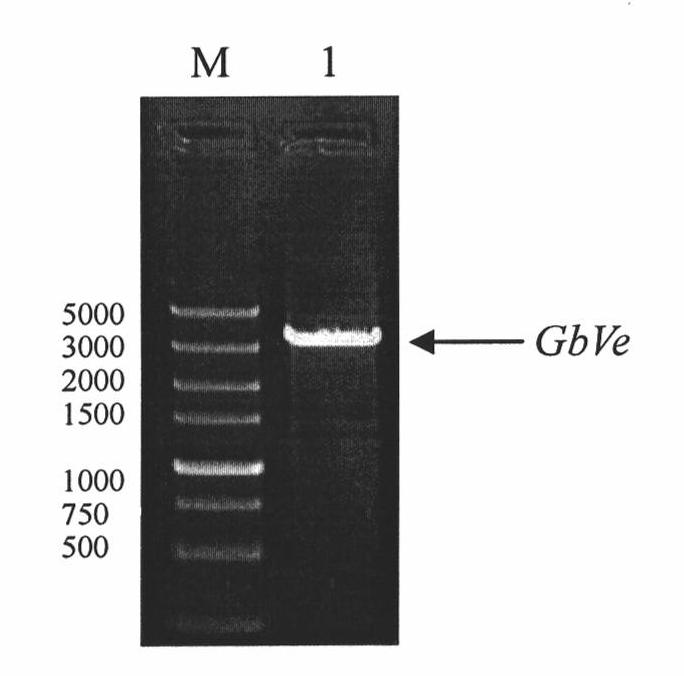
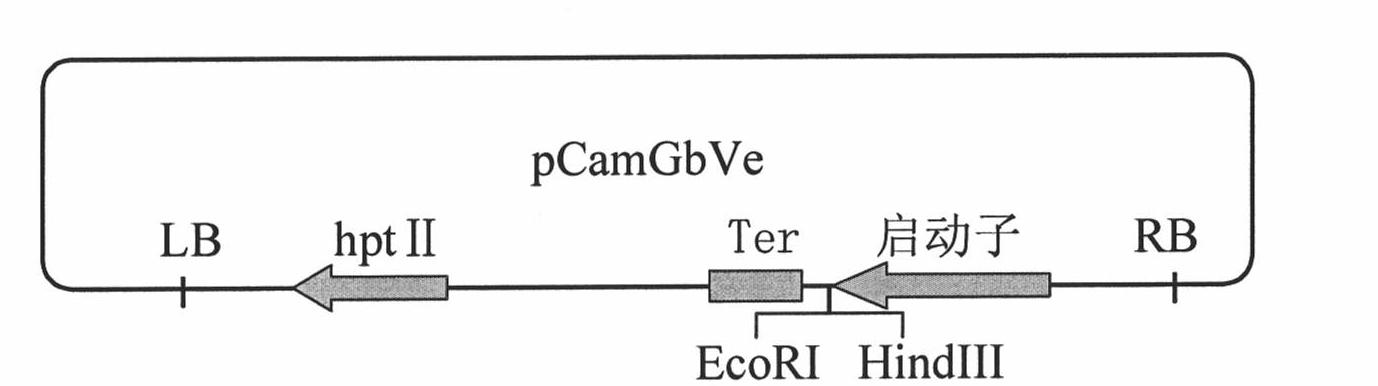
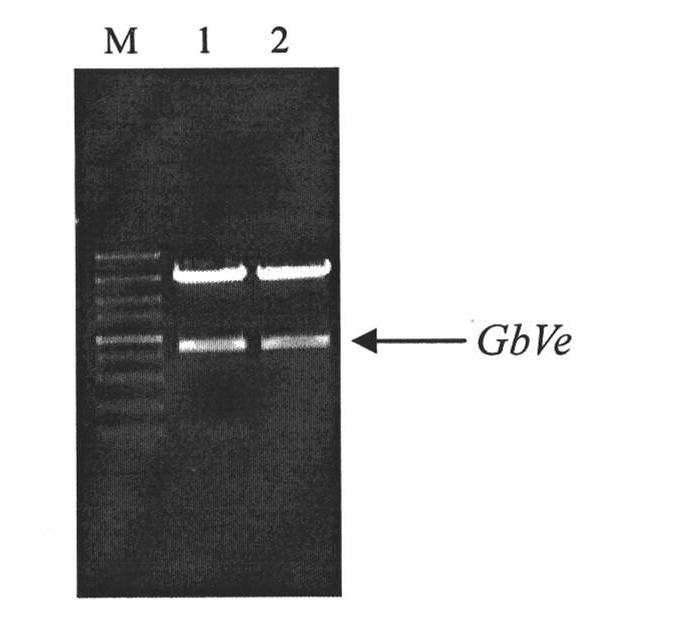
Cotton GbVe gene, protein coded thereby and use thereof in vegetable verticillium wilt resistance
InactiveCN101928338AVerticillium wilt resistance increasedHelps reveal molecular mechanismsFungiBacteriaBiotechnologyNucleotide
The invention relates to cotton GbVe gene and a protein coded thereby. In the invention, a Ve gene associated with verticillium wilt resistance is obtained by cloning in a gossypium barbadense variety having high verticillium wilt resistance; a 5'-terminal GbVe gene fragment and a 3'-terminal GbVe gene fragment, which are obtained by amplification, are spliced; a primer capable of amplifying the full-length GbVe gene is designed and synthesized according to a gene sequence; the sequence of the full-length GbVe gene is obtained by the conventional colony sequencing method according to an RT-PCR technique; and the nucleotide sequence of the full-length GbVe gene is represented by SEQ ID No.1. In the invention; a new and important candidate gen is provided for vegetable verticillium wilt resistant gene project; and the obtained full-length GbVe gene and a vegetable expression vector constructed by the full-length GbVe gene lay a foundation for further transforming important crops for improving the verticillium wilt resistance of transgenic crops and have great economic benefits and application values.
Owner:HEBEI AGRICULTURAL UNIV.
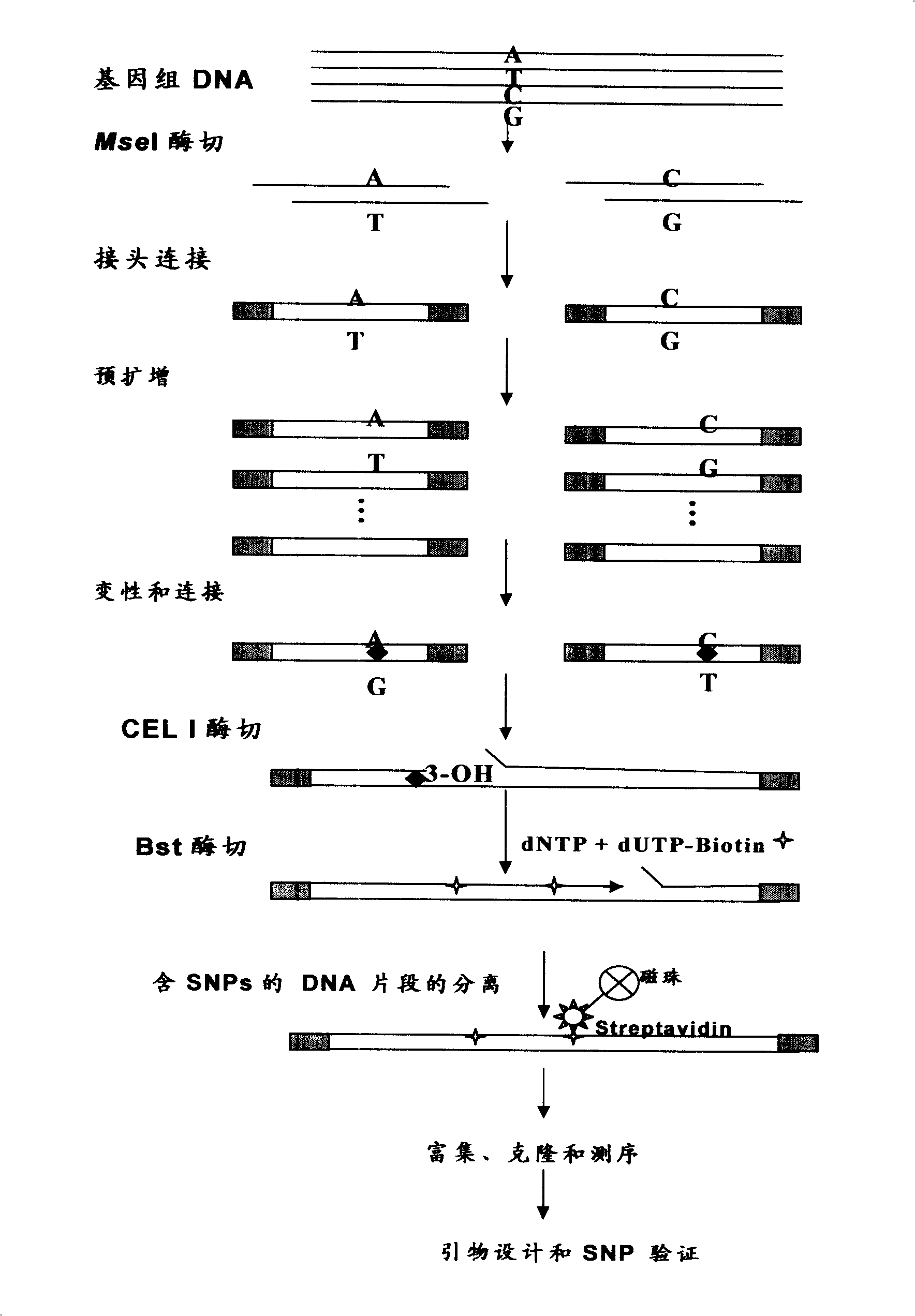
Aquatic product animal SNP mark screening method
InactiveCN101343667AEfficient separationImprove developmentMicrobiological testing/measurementAquatic animalGenetic diversity
The invention provides an aquatic animal SNP marking and selection method, which can solve the problems of low efficiency and high cost existing in the current SNP marking and selection technique. The method adopts the technical proposal comprising the following steps: firstly, genome DNA abstraction; secondly, MseI restrictive enzymes cutting, connection of connecting ends and pre-amplification; thirdly, heterozygous double chain forming, CEL I enzyme cutting and Bst DNA polymerase extending; fourthly, separation of beads containing SNP locus DNA molecule; fifthly, object DNA amplification, concentration and clone sequencing; and sixthly, SNP locus identification. The new SNP marking and selection method can efficiently, plentifully and randomly separate SNP under the condition of unknown genome information, thereby being favorable for the development and the subsequent utilization of the abundant SNP, and having significant application value and popularization prospect in aquatic animal SNP marking and selection, germplasm identification, genetic diversity evaluation, molecular breeding and the like.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Whole-genom sifting method for BPDE carcinogen related gene
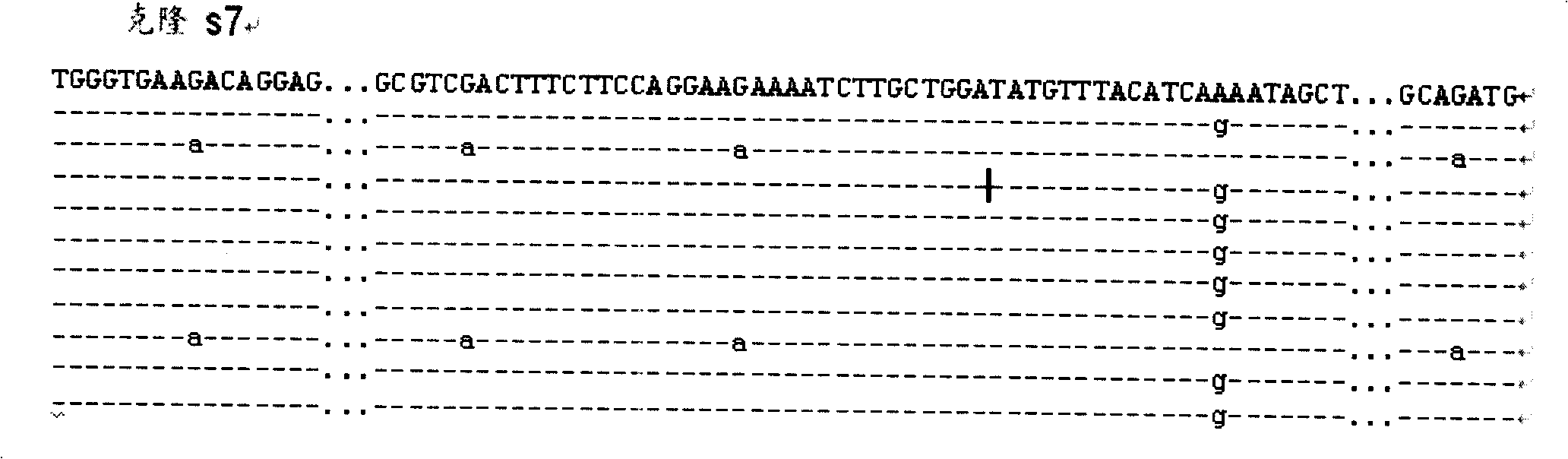
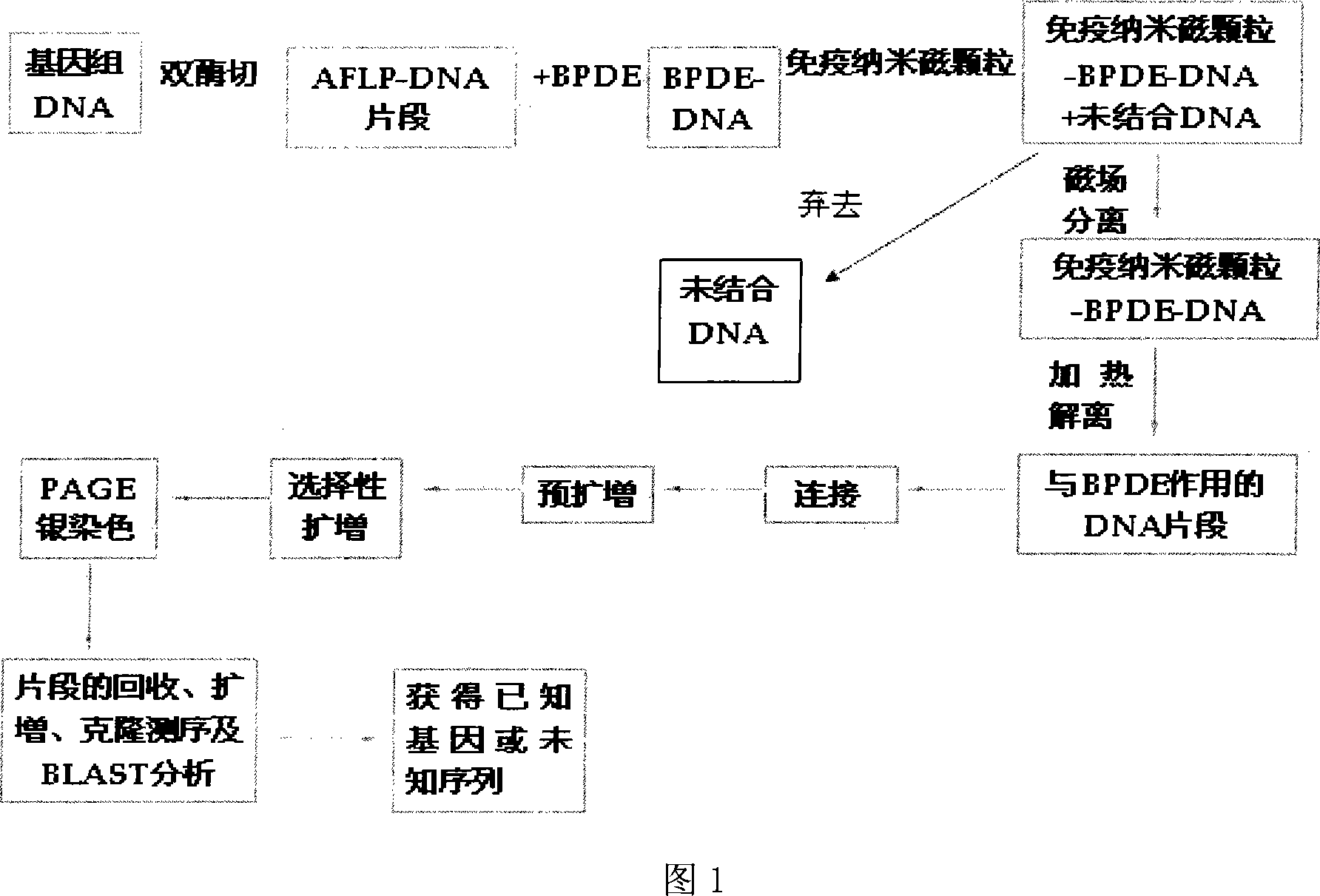
InactiveCN101113474AMicrobiological testing/measurementDNA/RNA fragmentationCarcinogenScreening method
The invention discloses a whole genome screening method for screening relative BPDE oncogenes and comprises the steps that: (1) genome DNA is extracted from normal lung tissue; (2) AFLP DNA fragment is prepared; (3) immunomagnetic of AFLP DNA fragment interacting with BPDE is enriched and separated; (4) AFLP PCR is increased; (5) denatured polyacrylamide gel electrophoresis and silver stain visualization are done to selective PCR amplification products of AFLP; (6) differential fragment is recycled, amplified, cloned and tested for sequence and homologous similarity and analysis are done. The whole genome screening method for screening relative BPDE oncogenes of the invention is the high effective and specific method and more BPDE susceptible oncogenes are screened in the whole genome level, which provides a basis for stating BPDE carcinogenic molecular mechanism.
Owner:INST OF HYGIENE & ENVIRONMENTAL MEDICINE PLA ACAD OF MILITARY MEDICAL
Multiple PCR detection primers for simultaneously detecting HPV, MBV and IHHNV of prawns, kit and application
ActiveCN105316330AGood repeatabilityImprove stabilityMicrobiological testing/measurementDNA/RNA fragmentationVirus strainCloning sequencing
The invention discloses multiple PCR detection primers for simultaneously detecting HPV, MBV and IHHNV of prawns, a kit and application. The primers are respectively HPV-F and HPV-R, MBV-F, MBV-R, IHHNV-F and IHHNV-R, sequences are shown as SEQ ID NO:1, SEQ ID NO:2, SEQ ID NO:3, SEQ ID NO:4, SEQ ID NO:5 and SEQ ID NO:6. The primers do not cross mutually, do not cross with other viruses, are low in false positive rate, can specifically detect out three different types of virus strains distributed around the world, has wide applicability, very high specificity and sensitivity and is low in detection limit. In addition, detected virus gene segments are not needed to undergo cloning, sequencing and sequence alignment. Whether a sample to be tested contains one, two or three of the three types of viruses or not can be detected out through one experiment.
Owner:THIRD INST OF OCEANOGRAPHY STATE OCEANIC ADMINISTATION
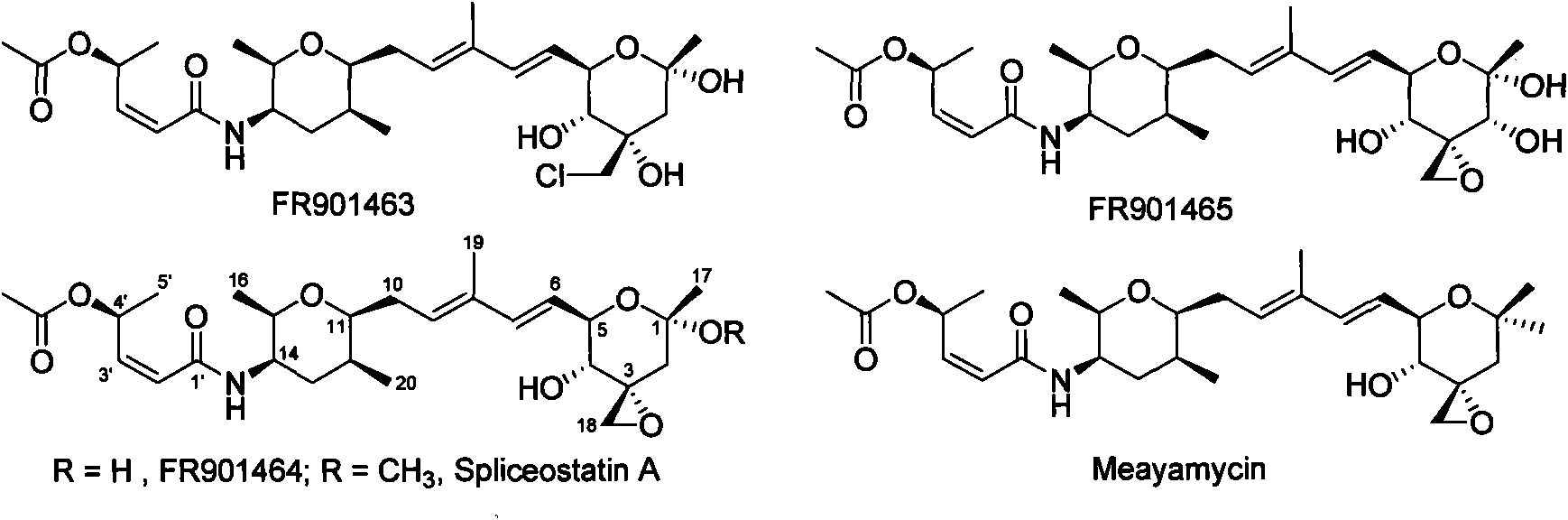
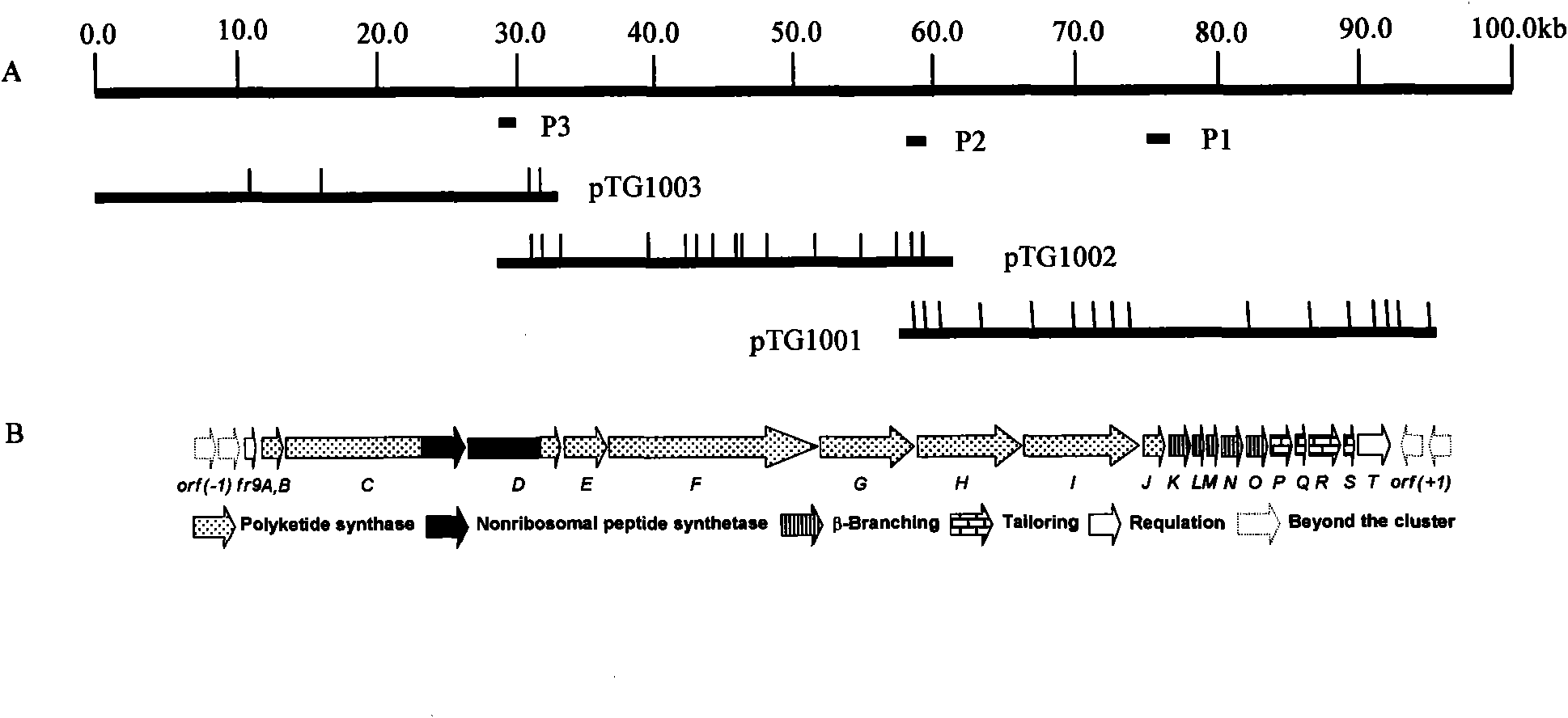
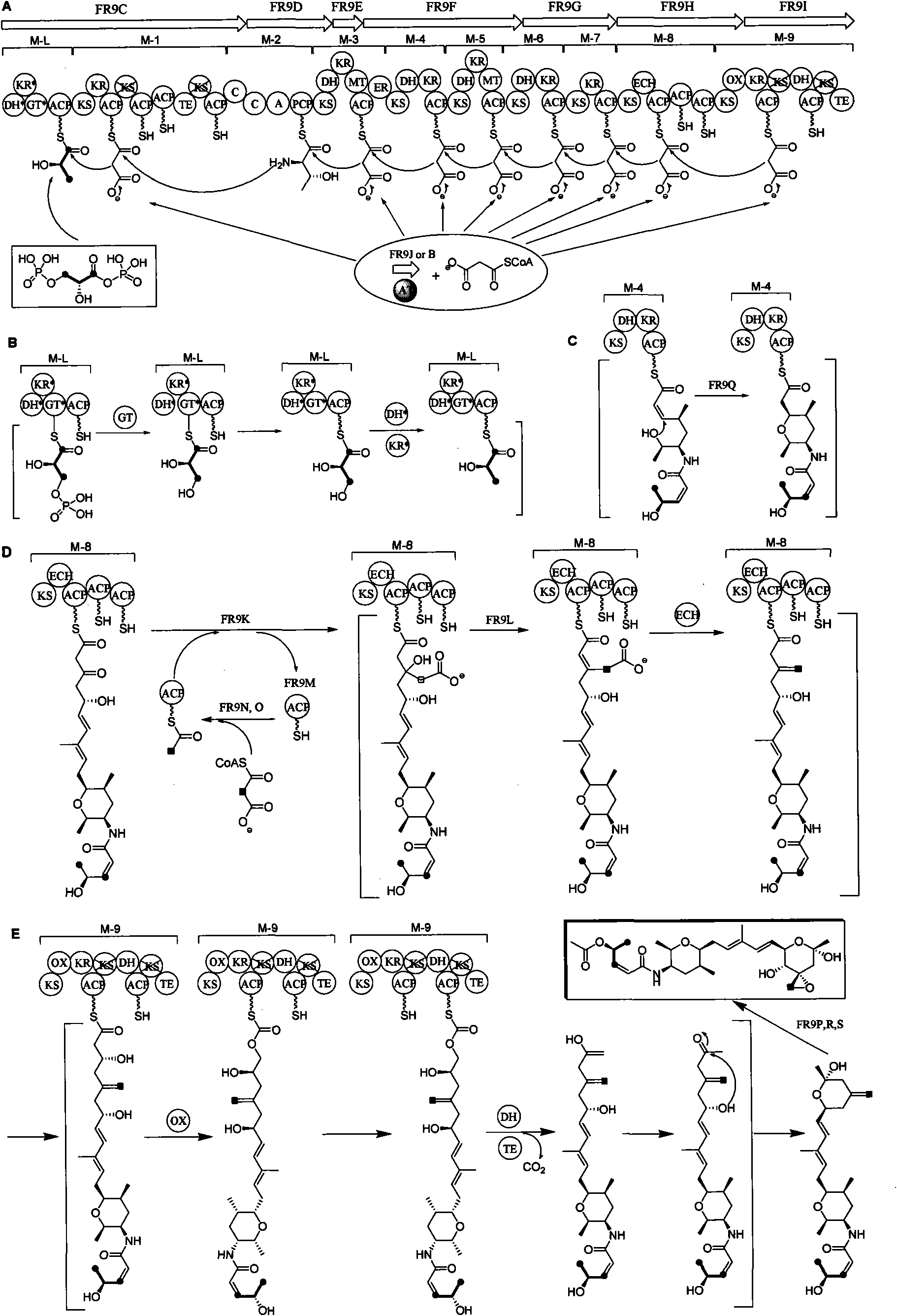
Biosynthetic gene cluster of FR901464
ActiveCN101818158AUnderstanding Biosynthetic MechanismsDepsipeptidesFermentationBiosynthetic genesGenetic engineering
The invention relates to cloning, sequencing, analysis and functional study of a biosynthetic gene cluster of a natural product FR901464 which has anti-tumor activity and is generated from pseudomonas as well as the application thereof. The whole gene cluster contains 20 genes: five polyketide synthase genes, one hybrid polyketide / non-ribosomal polypeptide synthase gene, one non-ribosomal polypeptide synthase gene, three independent acyltransferase genes, four genes related to polyketide backbone alkylation, four post-modification genes and two control-related genes. The genetic operation of the biosynthetic gene can block the biosynthesis of FR901464. The provided gene and protein thereof can be used for genetic engineering, protein expression, enzyme catalysis reaction, and the like of the compound and can also be used for searching and discovering compounds that can be used in medicines, industry or agriculture or genes and proteins thereof.
Owner:SHANGHAI INST OF ORGANIC CHEM CHINESE ACAD OF SCI
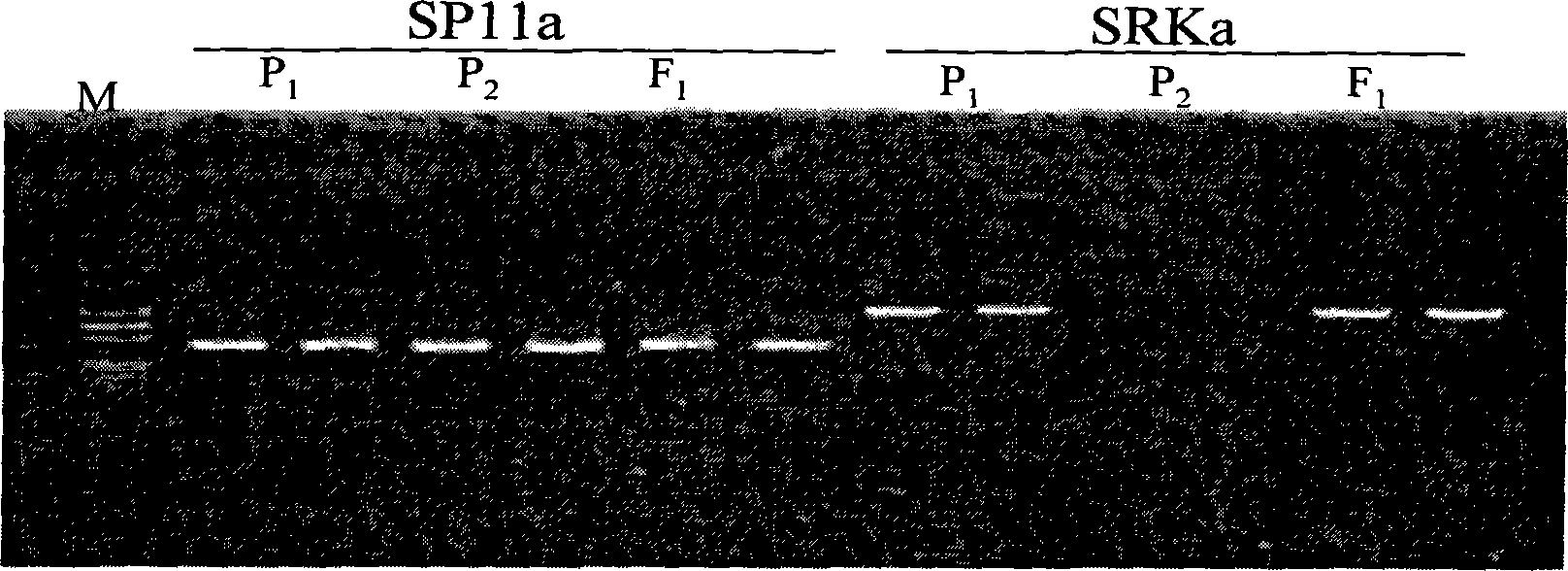
Molecule marker of brassica napus self-incompatible maintainer as well as preparation and uses thereof
InactiveCN101250524AReduce breeding workloadShorten the breeding periodMicrobiological testing/measurementDNA preparationBrassicaDNA fragmentation
The invention belongs to the rapeseed breeding technical field and in particular relates to a dominant SCAR molecular marker of a brassica napus self-incompatible maintainer line, which is characterized in that a reported primer SP11a is utilized to amplify genome DNA of a brassica napus self-incompatible line S-1300 and the brassica napus self-incompatible maintainer line, amplified DNA fragments are respectively obtained, then, the amplified DNA fragments are cloned and sequenced to compare nucleotide sequences, a primer SP11b is designed according to the difference of the sequences, PCR is amplified, the dominant SCAR molecular marker of the brassica napus self-incompatible maintainer line is obtained, and the nucleotide sequence of the dominant SCAR molecular marker is displayed in a sequence table SEQ ID NO:3. The invention also discloses a method for preparing the molecular marker and application of the molecular marker.
Owner:HUAZHONG AGRI UNIV
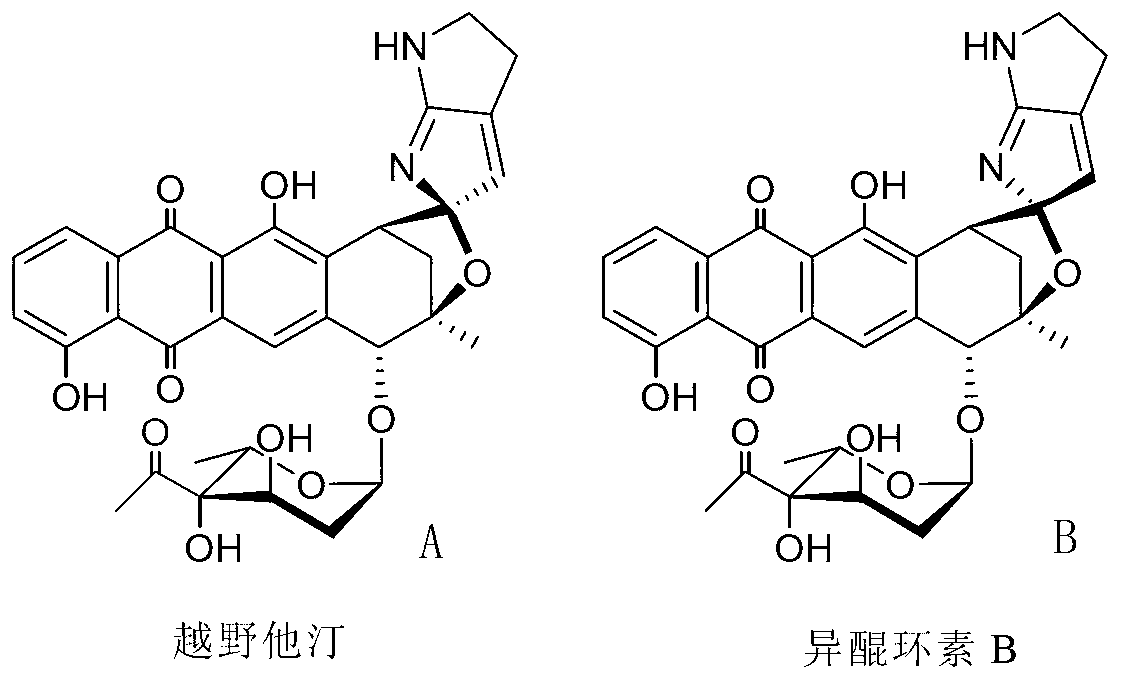
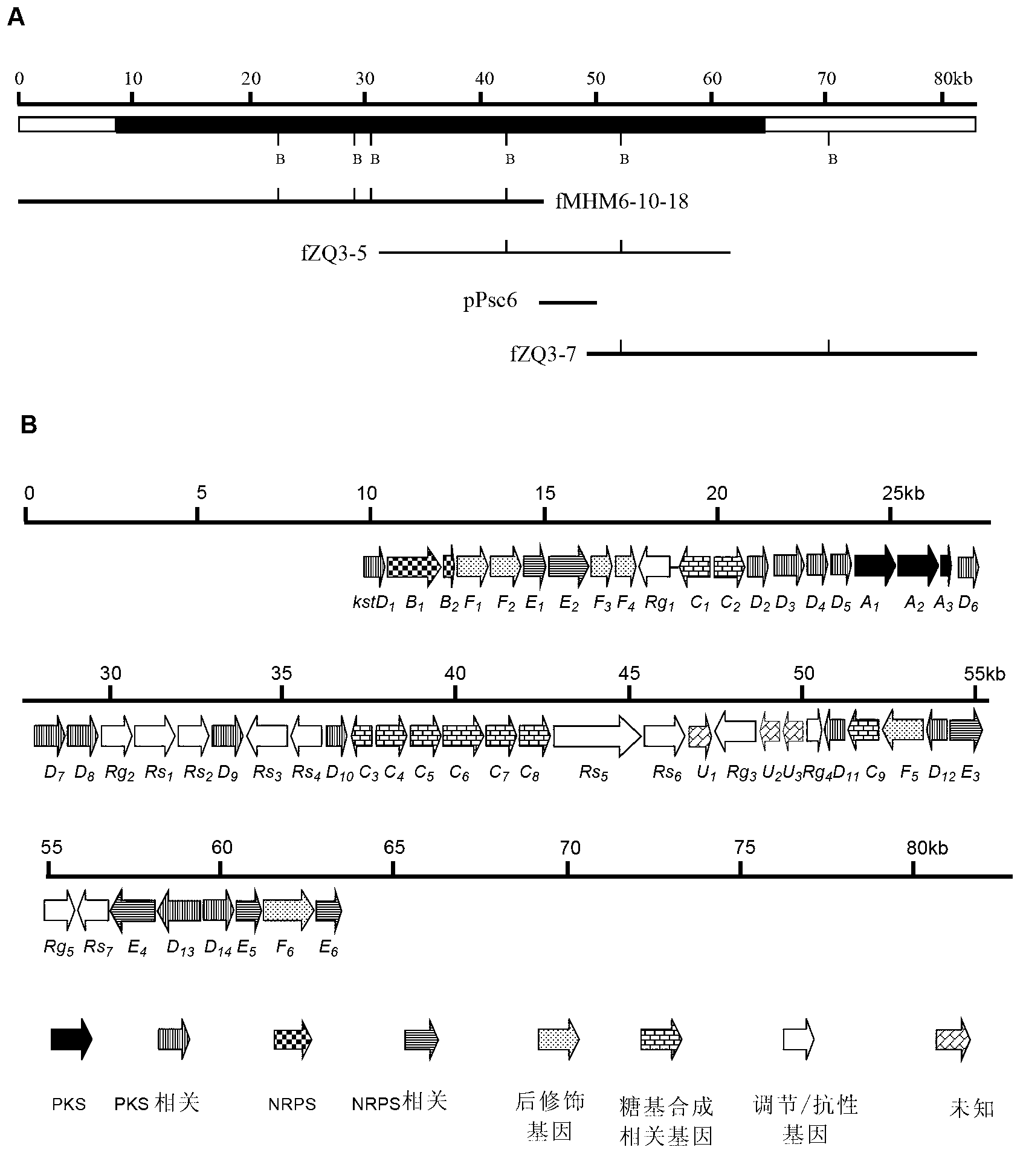
Biosynthesis gene cluster of kosinostatin and application thereof
The invention relates to a biosynthesis gene cluster of kosinostatin, and in particular relates to cloning, sequencing, analysis, functional study and application of the biosynthesis gene cluster of the kosinostatin, which is an antharcycline antibiotic with antitumor activity and generated from micromonospora sp.TP-A0468. The whole gene cluster comprises 55 genes in all, that is, 17 II-type polyketone synthetase (PKS) related genes, 8 non-ribosome polypeptide synthetase (NRPS) related genes, 9 glycosyl synthetic related genes, 6 special post-modifying genes, 7 antibiotic genes, 5 conditioning genes and 3 genes without definite functions. Through the genetic manipulation of the biosynthesis genes, the biosynthesis of kosinostatin can be blocked, the output is changed, or novel compounds are produced. The gene cluster can be applied to gene engineering, protein expression, enzymatic catalytic reaction and the like of antharcycline compounds, and can also be used for finding and discovering compounds, genes or proteins for medicines, industry or agriculture.
Owner:SHANGHAI INST OF ORGANIC CHEMISTRY - CHINESE ACAD OF SCI
Rice anti-Nilaparvata-lugens gene Bph28 and application thereof
ActiveCN104263737AObvious resistance to brown planthopperImproved performance against brown planthopperPlant peptidesFermentationBiotechnologyCandidate Gene Association Study
The invention discloses a rice anti-Nilaparvata-lugens gene Bph28 and application thereof. The rice anti-Nilaparvata-lugens gene Bph28 has a nucleotide sequence disclosed as SEQ.ID.NO.1. A biological informatic analysis and candidate gene clone sequencing method is utilized to separate the rice anti-Nilaparvata-lugens gene Bph28; and the Bph28 is subjected to genetic transformation, so that the insect-sensitive rice has anti-Nilaparvata-lugens phenotype, which proves the anti-Nilaparvata-lugens function of the gene.
Owner:CROP INST SICHUAN PROVINCE ACAD OF AGRI SCI
Aptamer screening method of no-fixed point target substance
ActiveCN103114147AEasy to fixEasy to operateMicrobiological testing/measurementBiotin-streptavidin complexScreening method
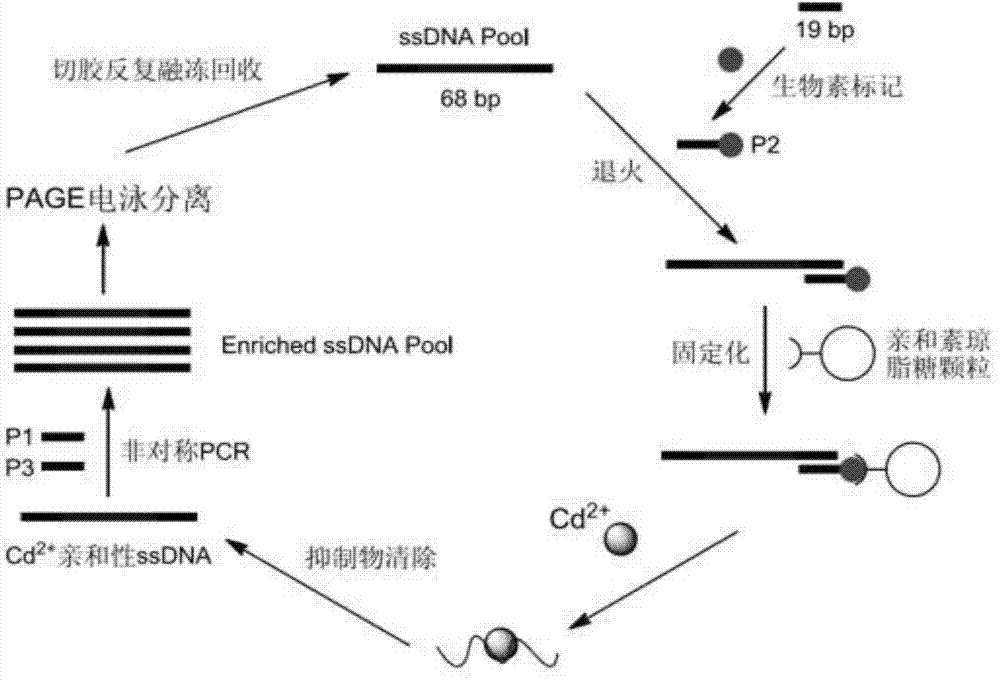
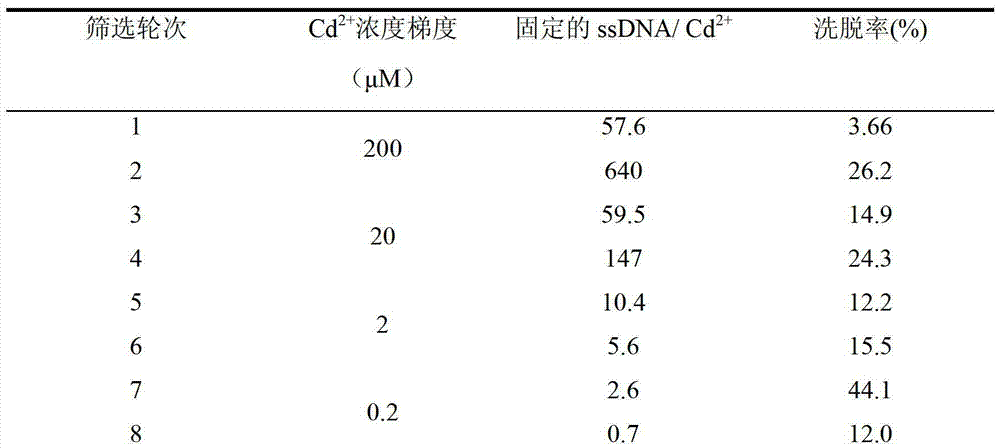
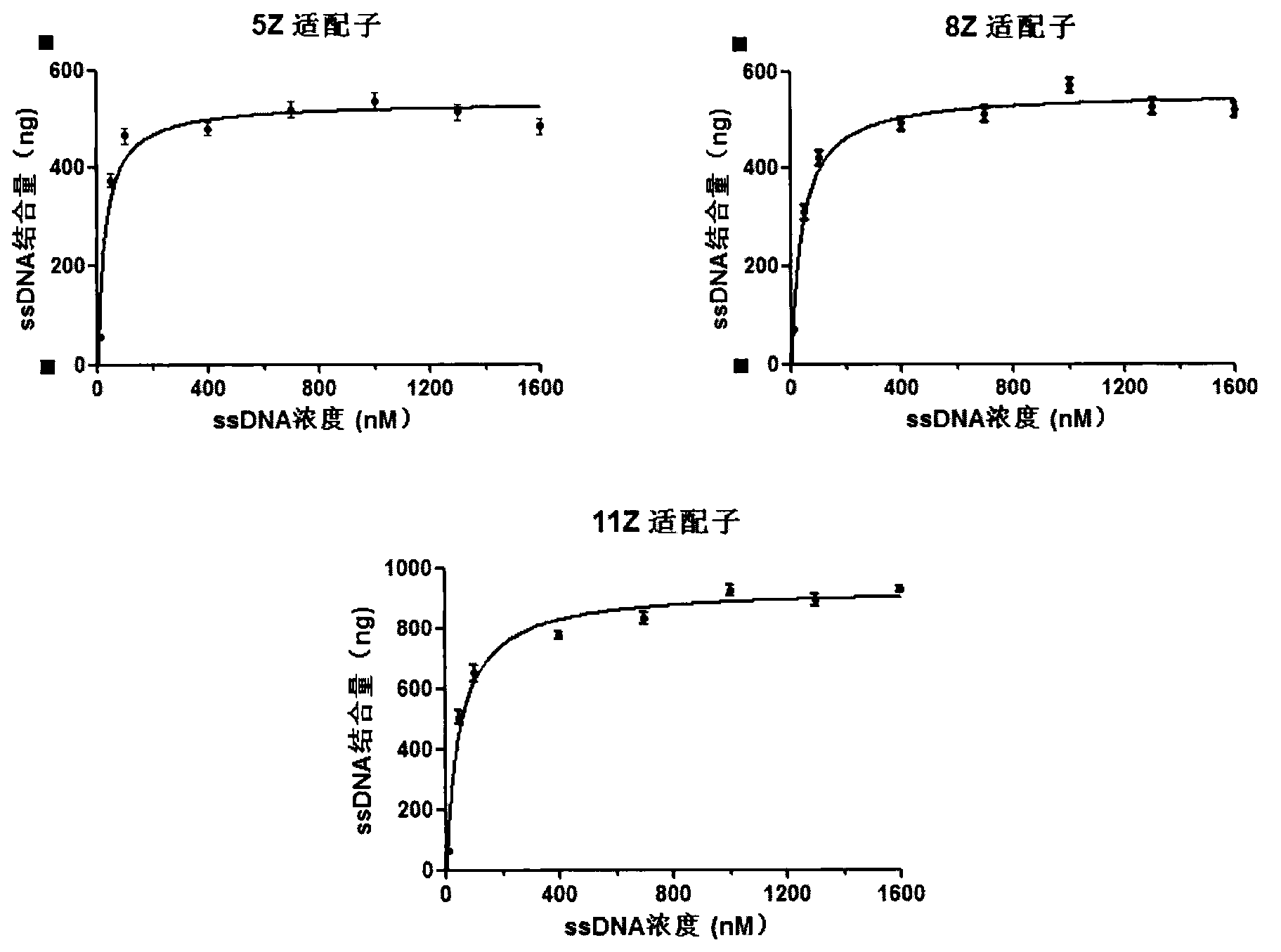
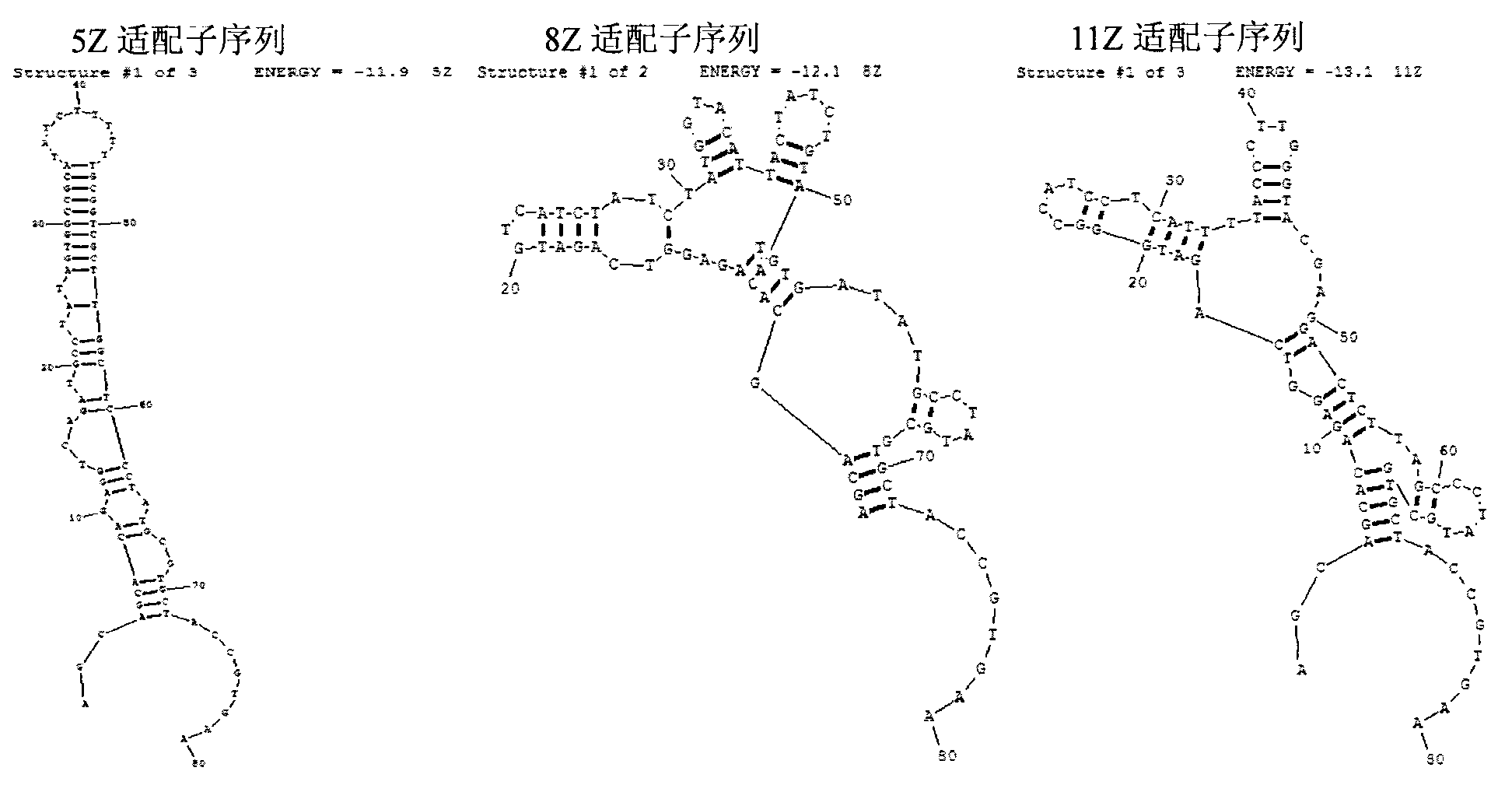
The invention discloses an aptamer screening method of a no-fixed point target substance in the technical field of nucleic acid aptamer screening in analytical chemistry. The method comprises the following steps of: screening in a systematic evolution of ligands by an exponential enrichment (SELEX) method by building a random oligonucleotides library, and an upstream primer, a downstream primer and a fixed oligonucleotides library primer for polymerase chain reaction (PCR) proliferation and utilizing an agarose particle fixed oligonucleotides library with a streptavidin mark to obtain a PCR product for clone sequencing; and obtaining aptamers bonded with the no-fixed point target substance with the special affinity after the sequencing is finished. The method is applied to screening of heavy metal cadmium ion aptamers, so that 13 aptamers which are bonded with the cadmium ion with the high specificity and high affinity are obtained; and therefore, the aptamer screening method is good in fixing effect, high in adaptability, low in cost and suitable for aptamer screening of the no-fixed point target substance.
Owner:SHANGHAI JIAO TONG UNIV
Oligonucleotides aptamer special for distinguishing zearalenone
ActiveCN103013998AHigh affinityStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationNucleotideMagnetic bead
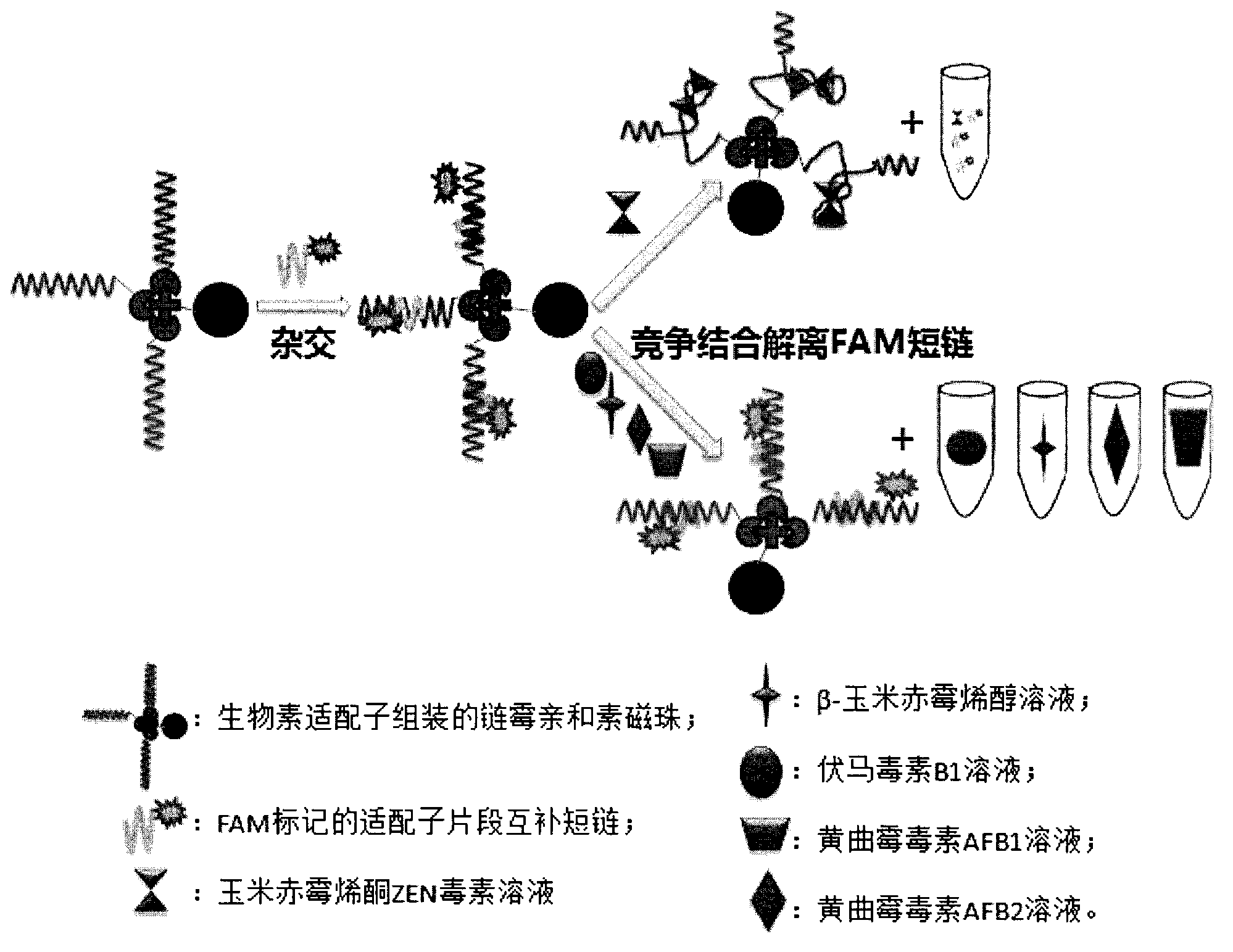
The invention discloses a group of oligonucleotides aptamers special for distinguishing zearalenone, and belongs to the field of food safety testing. An SELEX (systematic evolution of ligands by exponential enrichment) technology is combined with a magnetic bead with zearalenone coupled on the surface, and after 14 rounds of repeated incubation, cleaning, dissociation, amplification, gamma-exonuclease digestion are carried out to prepare a single-stranded secondary library, each oligonucleotides aptamer capable of being combined with the specificity of the zearalenone is screened out from a random single-stranded DNA library, after clone sequencing and 12 typical sequences are synthesized, the affinity is analyzed, and a specificity test is carried out to three aptamers with best affinity. The group of oligonucleotides aptamers (a nucleic acid sequence thereof selected from 1-3 in a sequence list) provides a new choice for developing a method which is for replacing the existing methods for detecting the zearalenone by depending on an antibody.
Owner:JIANGNAN UNIV
Preparation method for procambarus clarkia simple sequence repeats (SSR) primer
ActiveCN104726554AEasy to filterQuick filterMicrobiological testing/measurementDNA preparationProcambarusMagnetic bead
The invention discloses a preparation method for a procambarus clarkia simple sequence repeats (SSR) primer. The preparation method comprises the steps: (1), extracting procambarus clarkia genome DNAs: extracting the genome DNAs from procambarus clarkia abdominal muscles; (2), hybridizing a probe and a target segment: hybridizing the biotin labeled probe and the target segment to obtain a hybridization product; (3), enriching magnetic beads: enriching the hybridization product by a magnetic bead enrichment method to obtain an enrichment product; (4), performing positive clone sequencing, and designing a primer amplification gene DNA according to a microsatellite flanking sequence; (5), performing SSR primer detection. The preparation method for the procambarus clarkia SSR primer has the advantages that a microsatellite sequence of the procambarus clarkia whole genome can be simply and quickly screened; a large number of SSR primers can be developed at a short time; technical measures are provided for germ plasma conservation and genetic improvement of the procambarus clarkia.
Owner:ZHEJIANG INST OF FRESH WATER FISHERIES
Whole genome sequence of Seneca valley virus SVV/CH/ZZ/2016 and amplification primer thereof
The invention discloses a whole genome sequence of Seneca valley virus SVV / CH / ZZ / 2016 and an amplification primer thereof. The whole genome sequence of Seneca valley virus SVV / CH / ZZ / 2016 is obtained by the following steps: amplifying 7 nucleotide sequence fragments (S1, S2, S3, S4, S5, S6 and S7) of the strain of SVV / CH / ZZ / 2016 by a one-step RT-PCR method; performing clone sequencing on the 7 nucleotide sequence fragments; sequentially splicing, editing and correcting the DNA sequences of the 7 nucleotide sequence fragments, to finally obtain the whole genome sequence of Seneca valley virus SVV / CH / ZZ / 2016. The whole genome sequence of Seneca valley virus SVV / CH / ZZ / 2016 obtained in the invention is favorable for further studying the pathogenic mechanism, molecular epidemiology, reverse genetics and the like of the Seneca valley virus, and provides important data support and theoretical basis for the diagnostic reagent development, vaccine development and the like of the Seneca valley virus.
Owner:JINYUBAOLING BIO PHARMA CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com