Aptamer screening method of no-fixed point target substance
A screening method and unfixed technology, applied in the direction of biochemical equipment and methods, microbe determination/inspection, etc., to achieve the effect of strong applicability, convenient operation and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
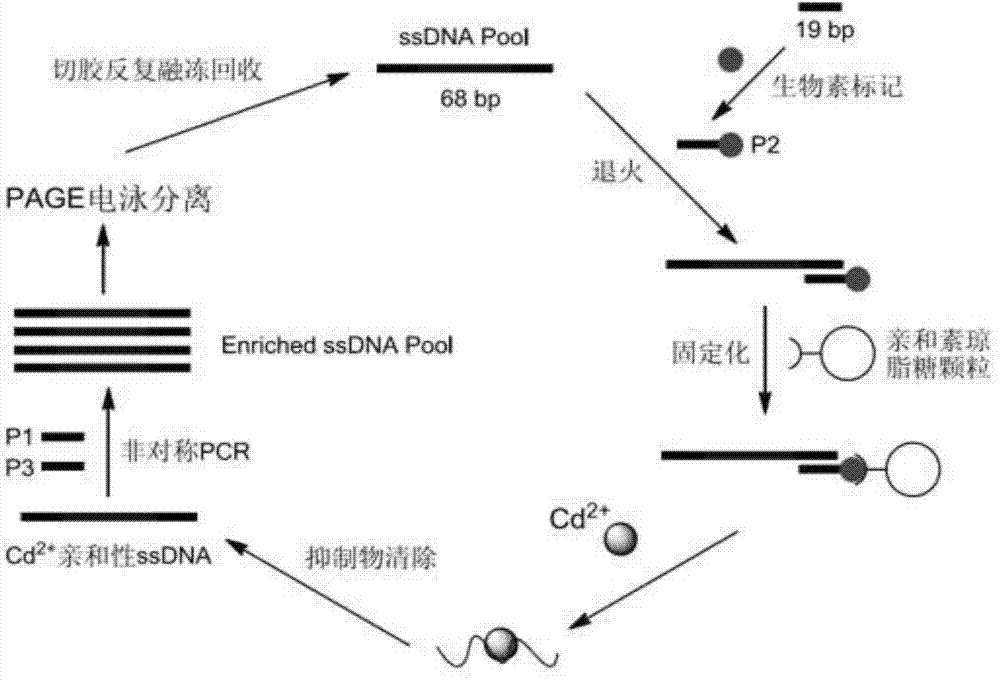
[0033] Such as figure 1 As shown, this embodiment includes the following steps:
[0034] Step 1) Construct a random oligonucleotide library with a total length of 68bp. The sequence of the oligonucleotide library is: 5'-ACCGACCGTGCTGGACTCT-N30-AGTATGAGCGAGCGTTGCG-3', wherein both ends are fixed sequences of 19bp, and the middle 30 bases The basis is a random sequence.
[0035] Step 2) design three primers, wherein P1 (5'-ACCGACCGTGCTGGACTCT-3') and P2 (5'-CGCAACGCTCGCTCATACT-3') are used as the upstream primer and downstream primer of PCR amplification oligonucleotide respectively, and P3 (5' -Biotin-CGCAACGCTCGCT CATACT-3') was used to immobilize the oligonucleotide library on streptavidin-labeled agarose particles.
[0036] Step 3) Take a random oligonucleotide library, mix it with the P3 sequence uniformly at a volume ratio of 1:2-3, and anneal the mixture. The annealing conditions are: denaturation at 94°C for 60 seconds, annealing at 59°C for 60 minutes, and 25°C Exten...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com