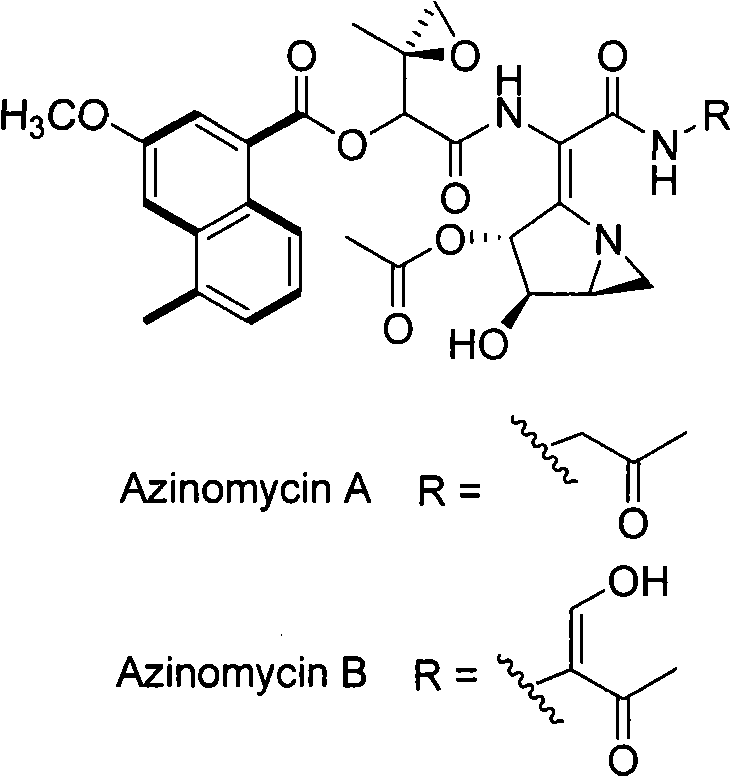
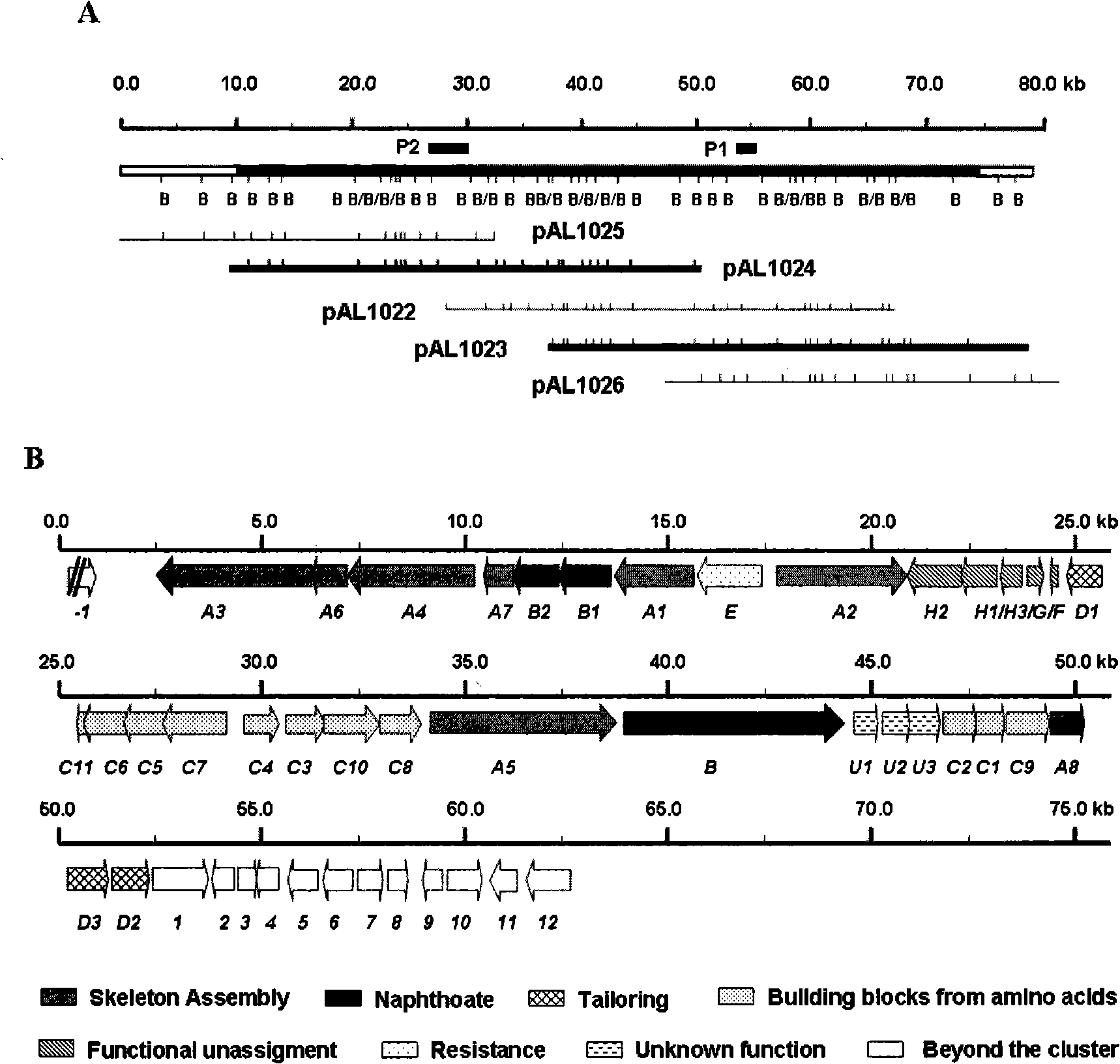
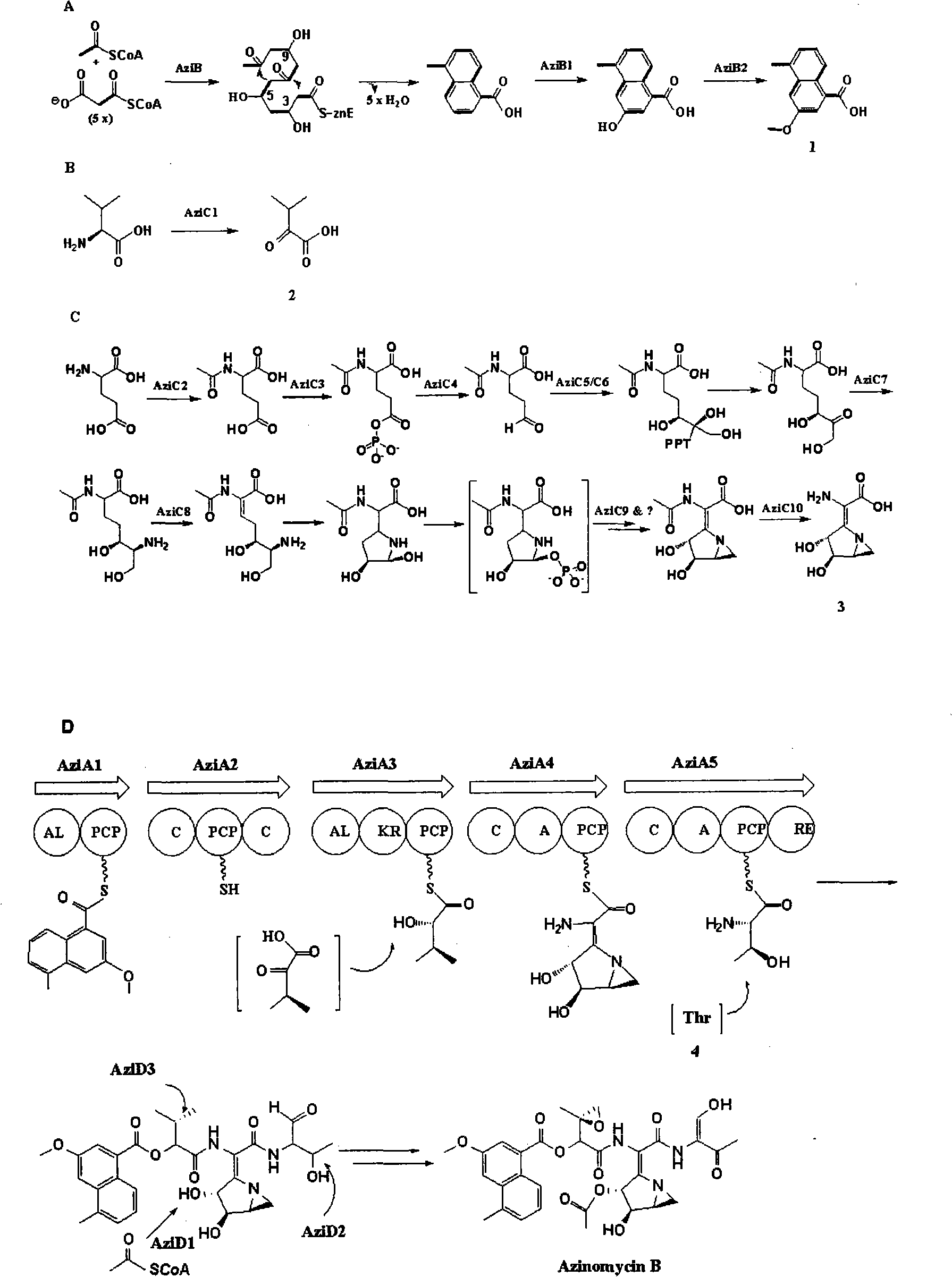
Biological synthesis gene cluster for Azintamide
A method of azithromycin and biosynthesis, which is applied in the field of microbial genetic resources and genetic engineering, and can solve the problems of high production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0101] Extraction of total DNA of azithromycin-producing bacteria Streptomyces sahachiroi NRRL2485:
[0102] 100 μL of 1 x 10 8 S.lavendulae 314 spore suspension was inoculated into 3mL TSB liquid medium, cultivated at 30°C, 230rpm for about 24hrs and reached the late logarithmic growth phase, then inoculated 2mL into 50mL TSB (containing 10mM magnesium chloride, 0.1% glycine), 30°C, After culturing at 250rpm for about 23hrs, it reached the early stage of the stable growth period, and it was milky yellow and turbid. The bacterial solution was centrifuged at 4°C and 3500rpm for 15min to collect mycelium, washed with lysate, and 0.5mL of pale milky yellow mycelia was collected. Add 10 mL of lysate (containing 5 mg / mL of lysozyme) to 1 mL of mycelium, a total of four tubes, vortex until uniform, and bathe in 37 ° C for 15 min. Add 0.1mL proteinase K (10mg / mL, freshly prepared with lysate), 1mL 10% SDS, mix well and quickly put in 70°C water bath for 15mim, it becomes clear. Coo...
Embodiment 2
[0104] Establishment of azithromycin-producing strain Streptomyces sahachiroi NRRL2485 genetic transfer system:
[0105] Culture E.coli S17-1 containing appropriate plasmids to OD 600 0.3-0.4, the bacterial cells in 20mL LB culture medium were collected by centrifugation, washed twice with an equal volume of LB, resuspended in 2mL LB, and used as E. coli donor cells. Take 500 μL of 20% glycerol spore suspension of Streptomyces sahachiroi NRRL2485 frozen at -80°C, wash twice with an equal volume of TES buffer (50 mM TES Na, pH 8.0), resuspend in an equal volume of TES buffer, Heat shock at 50°C for 10 min to germinate the spores. Add an equal volume of TSB medium and incubate at 37°C for 2-5hr. Centrifuge and resuspend in 0.5-1 mL LB as Streptomyces recipient cells. Mix 100 μL of different concentrations of recipient cells with an equal volume of donor cells and directly spread it on a medium containing 10 mM MgCl 2 After incubating at 30°C for 20 hours, gently wash the sur...
Embodiment 3
[0108] Construction of azithromycin-producing bacteria Streptomyces sahachiroi NRRL2485 genome library:
[0109] First, through a series of dilution experiments to determine the amount of Sau 3AI, on this basis, a large number of enzyme-digested DNA fragments slightly larger than 40kb, dephosphorylated. pOJ446 was first cut and dephosphorylated from the middle of the two cos sequences with Hpa I, and then cut with BamHI from the multiple cloning site to obtain two arms, which were ligated with the prepared 40kb DNA fragment overnight. Thaw the Promega Packagene extract taken out at -80°C on ice, add 10ul of the ligation product immediately, flick and mix well, and place it at room temperature (about 22°C) for 3hr. Add 445ul Phagebuffer, invert to mix; add 25ul chloroform to terminate the reaction, centrifuge to make the chloroform sink to the bottom, and store at 4°C. The strain E.coli LE392 frozen at -80°C was spread on LB medium for recovery. Take a single colony and inocu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com