Aquatic product animal SNP mark screening method
A screening method and technology for aquatic animals, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of low efficiency and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Taking semi-smooth tongue sole as an example below, the technical contents of the present invention are elaborated in conjunction with the accompanying drawings:
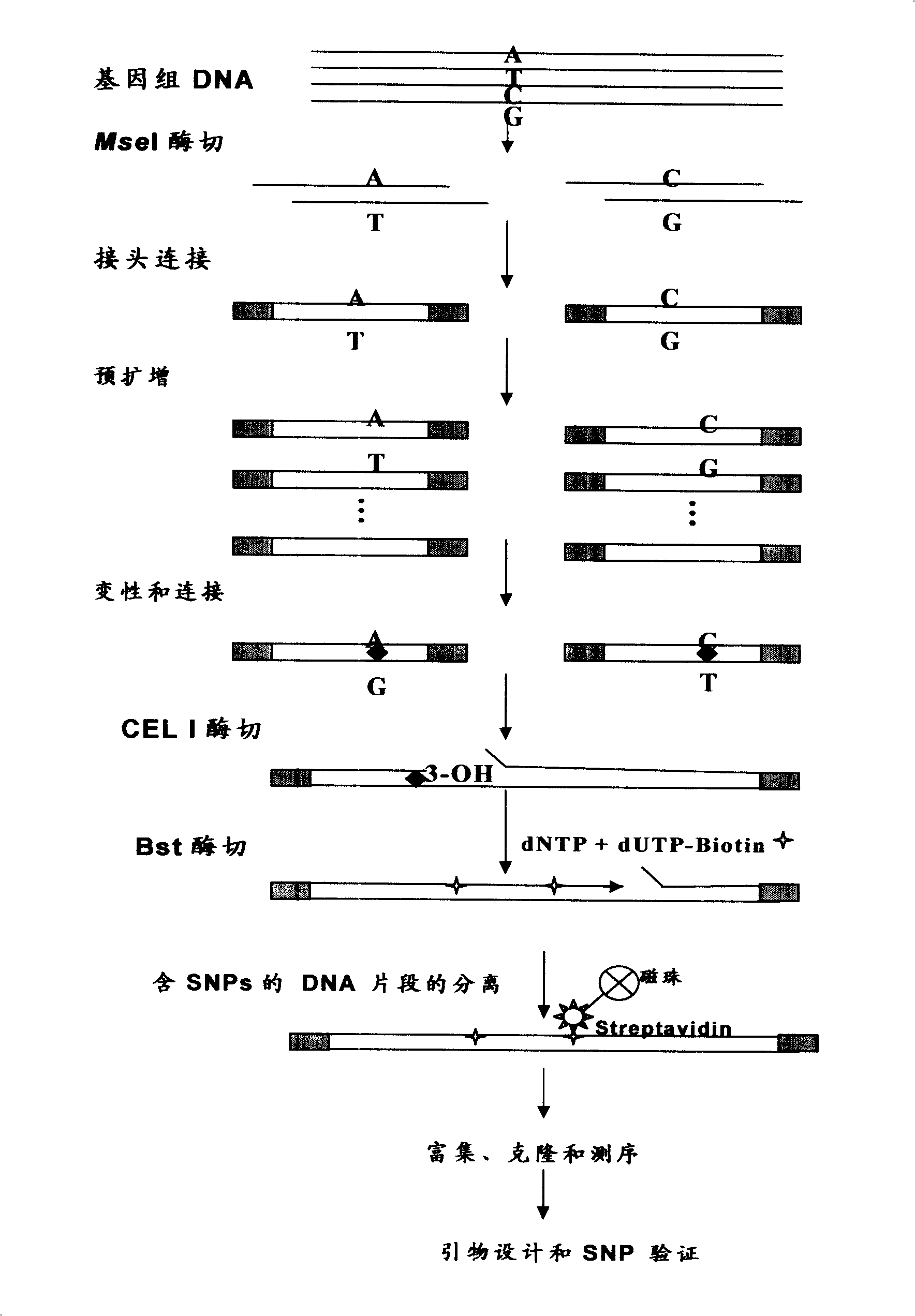
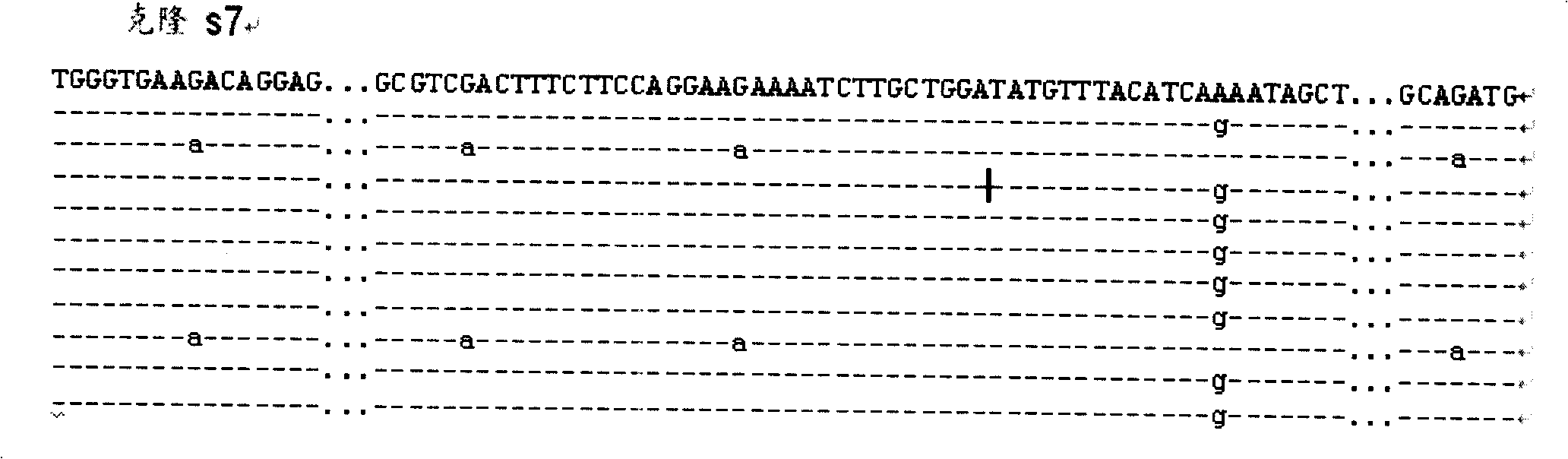
[0040] Such as figure 1 As shown, it includes six steps: 1) genomic DNA extraction; 2) MseI restriction endonuclease digestion, adapter ligation and pre-amplification; 3) hybrid duplex formation, CEL I digestion and Bst DNA polymerase Extension; 4) Magnetic bead separation of DNA molecules containing SNP sites; 5) Amplification and enrichment of target DNA and cloning sequencing; 6) Identification of SNP markers. in,
[0041] 1) Genomic DNA extraction
[0042] The DNA of 10 individuals of tongue sole was extracted by phenol-chloroform method. Take 10-20 mg of liver tissue and put it into a mortar, add a little liquid nitrogen and grind until the tissue is ground into powder. Add 1ml of DNA extraction solution TENS (10mM Tris-HCl pH 8.0, 100mM NaCl, 25mM EDTA, 0.5% SDS) for homogenization. Add an equal vo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com