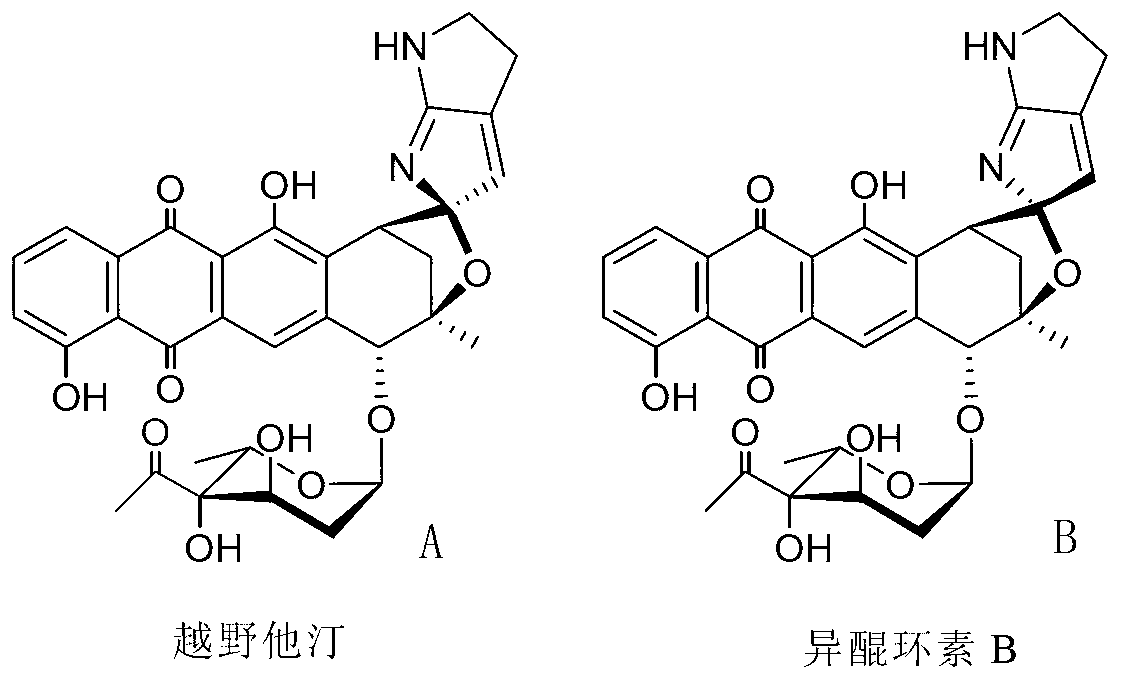
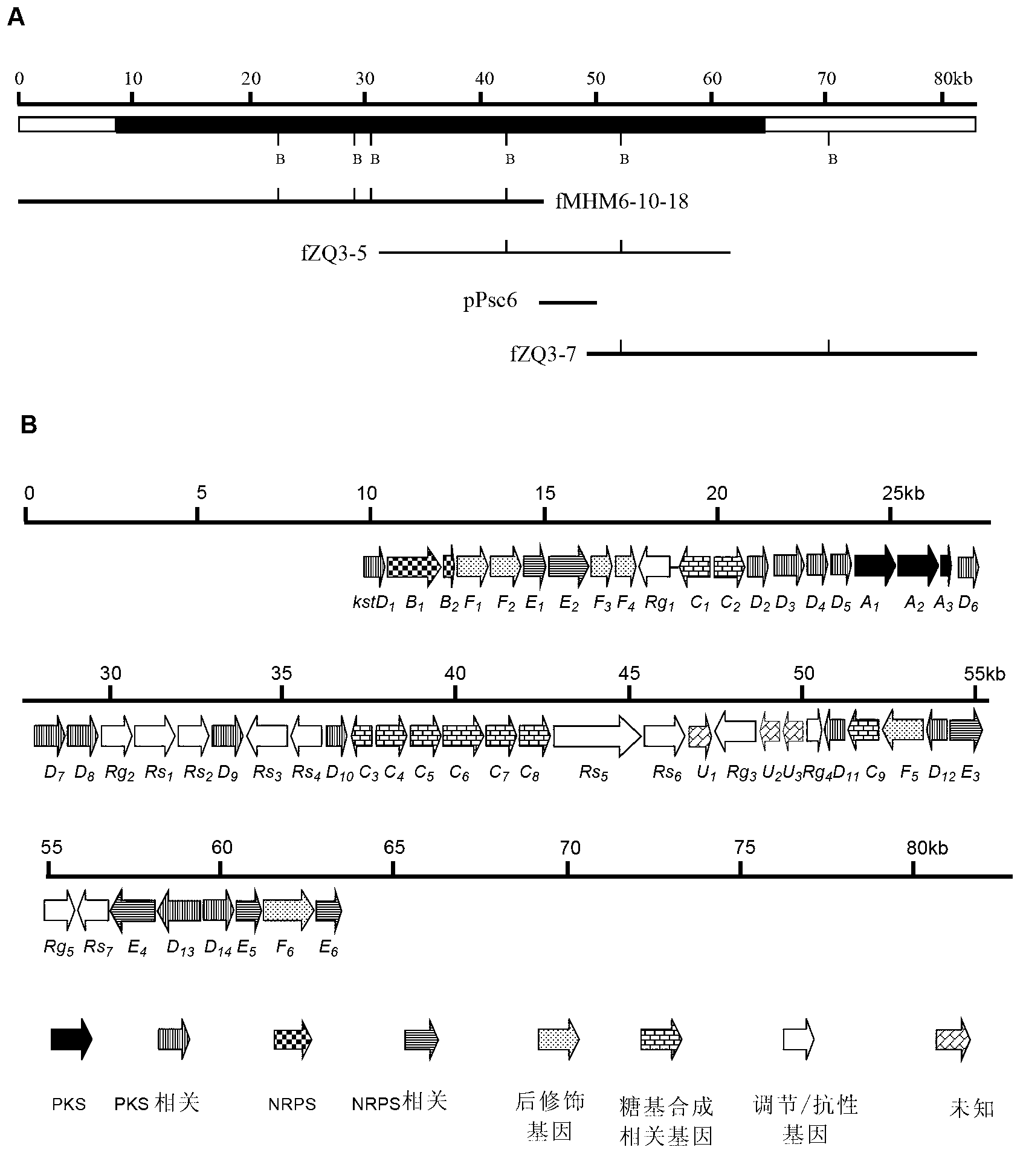
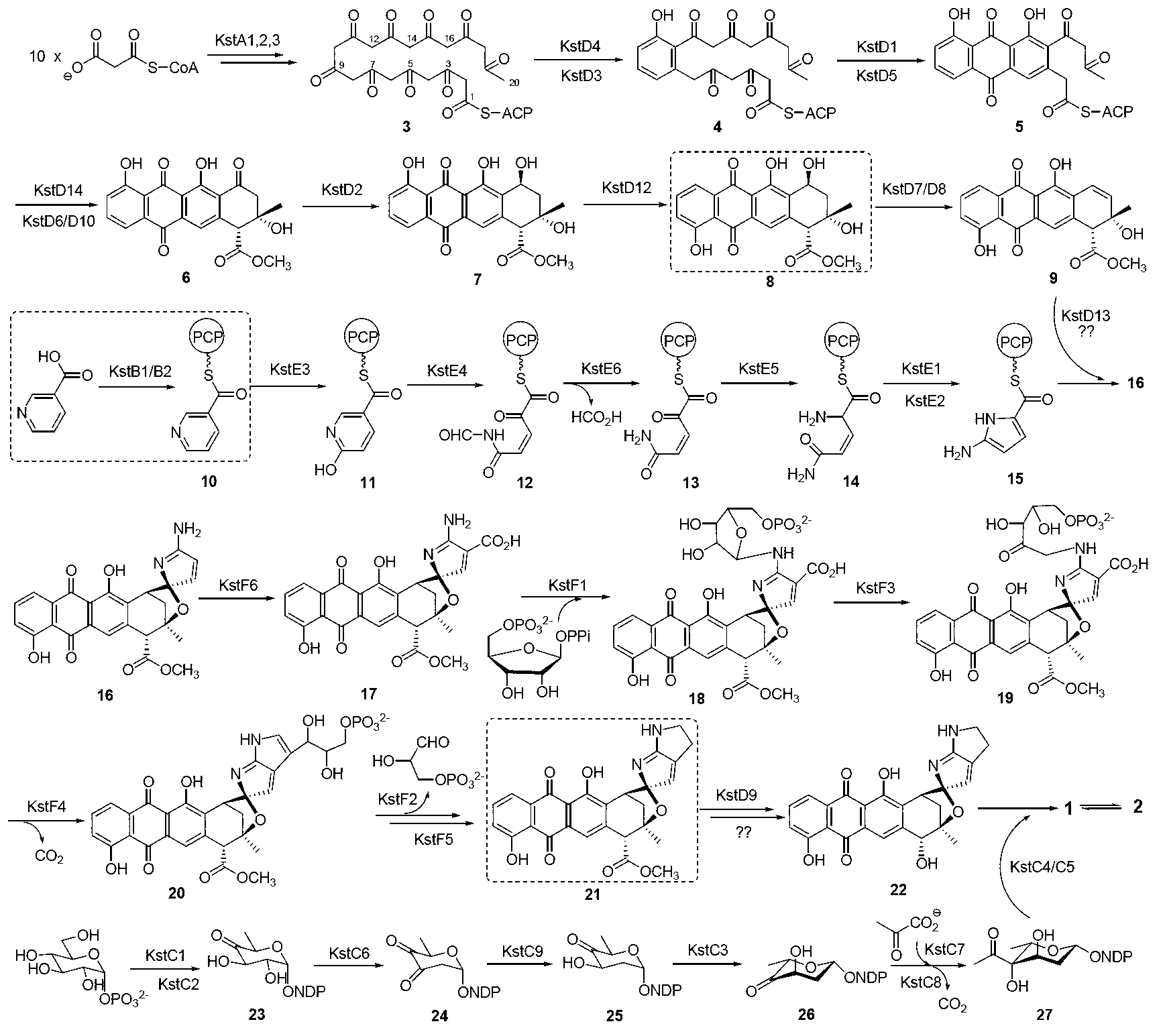Biosynthesis gene cluster of kosinostatin and application thereof
A biosynthesis and gene cluster technology, applied in the field of microbial gene resources and genetic engineering, can solve problems such as lack of understanding of synthetic pathways
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0202]Example 1 Extraction of Genomic DNA of Micromonospora sp.TP-A0468 Off-road Statin-producing Bacteria
[0203] 1) Collect mycelia
[0204] 100 μL 1 x 10 8 The spore suspension of Micromonospora sp.TP-A0468 reported by the Japanese scientist Tamotsu Furumai per mL was inoculated into 3 mL of ISP-2 liquid medium, cultivated at 30°C and 230 rpm for about 24 hours and reached the late logarithmic growth phase, and 2 mL was inoculated into In 50mL ISP-2 (containing 25mM magnesium chloride), culture at 30°C, 250rpm for about 23hrs and reach the early stage of the stable growth phase, which is milky yellow and turbid. Centrifuge the bacterial solution at 4°C, 3500rpm for 15min to collect mycelium, wash with lysis buffer, Collect 0.5 mL of pale milky yellow hyphae.
[0205] 2) Genomic DNA extraction
[0206] Add 10 mL of lysis buffer (containing 5 mg / mL lysozyme) to 1 mL of mycelia, vortex until uniform, and bathe in 37 °C water bath for 15 min. Add 0.1 mL of proteinase K sol...
Embodiment 2
[0207] Example 2PCR cloning off-road statin biosynthesis gene
[0208] The composition of the 50 μL PCR system is shown in Table 3:
[0209] table 3
[0210]
[0211]
[0212] Primers are shown in Table 4:
[0213] Table 4
[0214]
[0215]
[0216] PCR program:
[0217] Taq enzyme
[0218] Cycle 1: 94°C, 3min, 1 cycle;
[0219] Cycle 2: 94°C, 30s; 55-65°C, 30s; 72°C, 1min / kb; 35 cycles;
[0220] Cycle 3: 72°C, 5-10 min; 1 cycle.
[0221] Primestar:
[0222] Step 1: 98°C, 10s
[0223] Step 2: 58-65°C, 15s
[0224] Step 3: 72°C, 1min-3min
[0225] Repeat steps 1-3 for 30 rounds
[0226] Step 4: 72°C, 10min
[0227] The PCR conditions of different primers were optimized based on the above conditions.
[0228] After PCR, the DNA band of the expected size was recovered and purified by low-melting point gel, and connected to the PCR cloning vector pGEM-T Easy vector, transformed into E.coli DH5α, and coated on a medium containing ampicillin (ampicillin, A...
Embodiment 3
[0230] Example 3 Nucleic Acid Molecular Hybridization
[0231] Nucleic acid molecular hybridization:
[0232] 1) Digoxigenin DNA labeling: Dilute the DNA to be labeled with sterile water to a total volume of 15 μL, heat and denature it in a boiling water bath for 10 minutes, and immediately place it in an ice-salt bath to cool. Then add 2 μL of primer mixture, 2 μL of dNTP mixture, and 1 μL of enzyme, mix well, and place in a 37° C. water bath for about 16 hours. Add 0.8 μL of 0.8M EDTA (pH8.0) to terminate the reaction, add 2.5 μL of 4M LiCl and mix well, then add 75 μL of pre-cooled absolute ethanol to precipitate the labeled DNA, and place it at -80°C for 40 minutes. DNA was collected by centrifugation at 12000 rpm at 4°C for 20 minutes, the DNA pellet was washed with pre-cooled 70% ethanol, dried in vacuo and redissolved in 50 μL TE ((pH 8.0).
[0233] 2) Membrane transfer of colony hybridization (library screening): slightly thaw the gene library stored at -80°C, take 5...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com