DNA identification method of Isodon serra(Maxim.)Kudo and Rabdosia lophanthoides (Buch.-Ham. ex D. Don) Hara var. graciliflora (Benth.) Hara
A technology of Fragrant Tea Vegetables and Xihuangcao, which is applied in the field of plant species identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
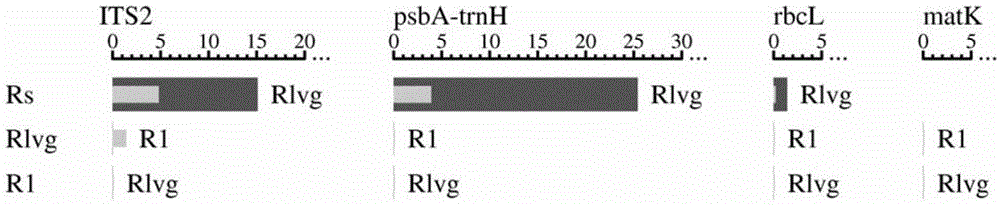
[0058] Example 1 Screening of DNA barcodes and establishment of a gene bank including rbcL gene sequence fragments
[0059] In this example, with regard to the screening of the DNA barcode genes of Xihuangcao, Xianwen Xiangchacai and Xianwen Xiangchacai (variety Xianhuaxiangchacai), the most suitable ones were selected from the 4 sequences of ITS2, matK, psbA-trnH, and rbcL. The DNA barcodes for the identification of Xihuangcao and Xianwenxiangchacai, that is, rbcL gene sequence fragments. In addition, this example established a gene bank including the rbcL gene sequence fragments of C. chinensis and C. chinensis.
[0060] 1 Instruments, materials and reagents
[0061] 1.1 Materials
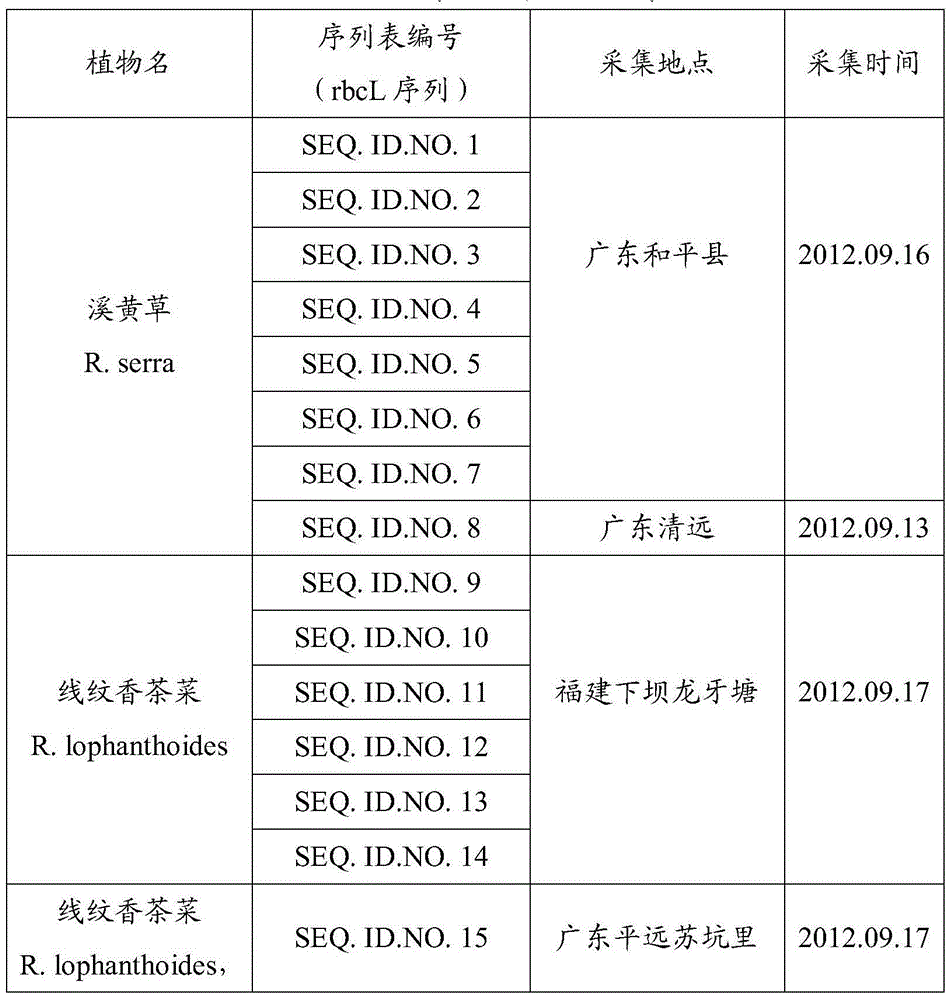
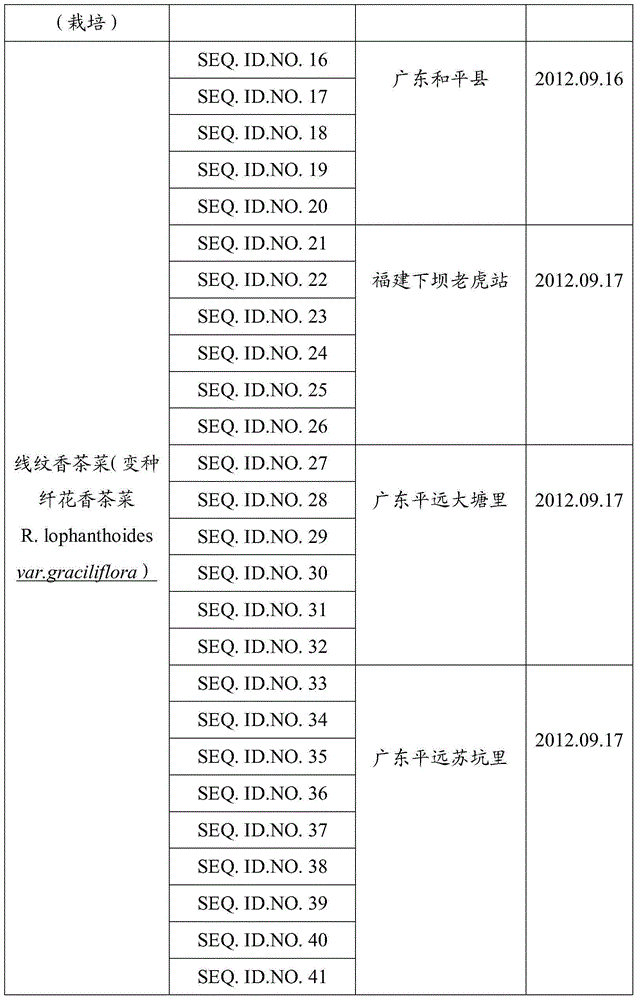
[0062] At 7 sampling points (Guangdong, Fujian), a total of 41 samples of Xihuangcao, Xianwen Xiangchacai and Xianwen Xiangchacai (variety Xianhuaxiangchacai) were collected, and the details are shown in Table 1 below. The following barcode screening experiments use plant leaves as materials...
Embodiment 2
[0104] Example 2 Identification of the Plant Species of Unknown Creek Yellow Grass and Streakia japonica
[0105] In this example, the method provided by the present invention is used to identify the samples of Unknown Xihuangcao and Cercis chinensis plant species.
[0106] Select the suspected but unidentified, fresh or dried whole grass of Xihuangcao and Xianwen Xiangchacai, and divide them into two parts, one part is identified by traditional taxonomic methods. Another copy was identified using the method used in the present invention.
[0107] Sample preparation for the method used in the present invention: unless otherwise specified, the surface of the medicinal material is scrubbed with 75% ethanol aqueous solution and then dried, and 5 mg to 2 g of leaves are weighed for later use.
[0108] The steps described in Example 1 were used to extract genomic DNA from leaves of plant tissues, and the rbcL gene sequence fragment was amplified using SEQ ID NO.42 and SEQ ID NO....
Embodiment 3
[0115] Example 3 Identification of Species of Unknown River Huangcao and Xianwen Xiangchacai Decoction Pieces
[0116] In this example, the method provided by the present invention is used to identify the species samples of Unknown Xihuangcao and Xianwen Xiangchacai decoction pieces.
[0117] The suspected but unidentified decoction pieces of Xihuangcao and Xianwen Xiangchacai were selected and divided into two parts, one part was identified by traditional taxonomic methods. Another part is identified by the method used in the present invention, and the identification steps are the same as in Example 2.
[0118] After identification by this method, the conclusion obtained is consistent with that of the traditional method.
[0119] However, after comparison, the requirements of this method for samples are easier to obtain than those of traditional methods.
[0120] The sample required by the method is a part of the plant where plant DNA is easily obtained. Although the deco...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com