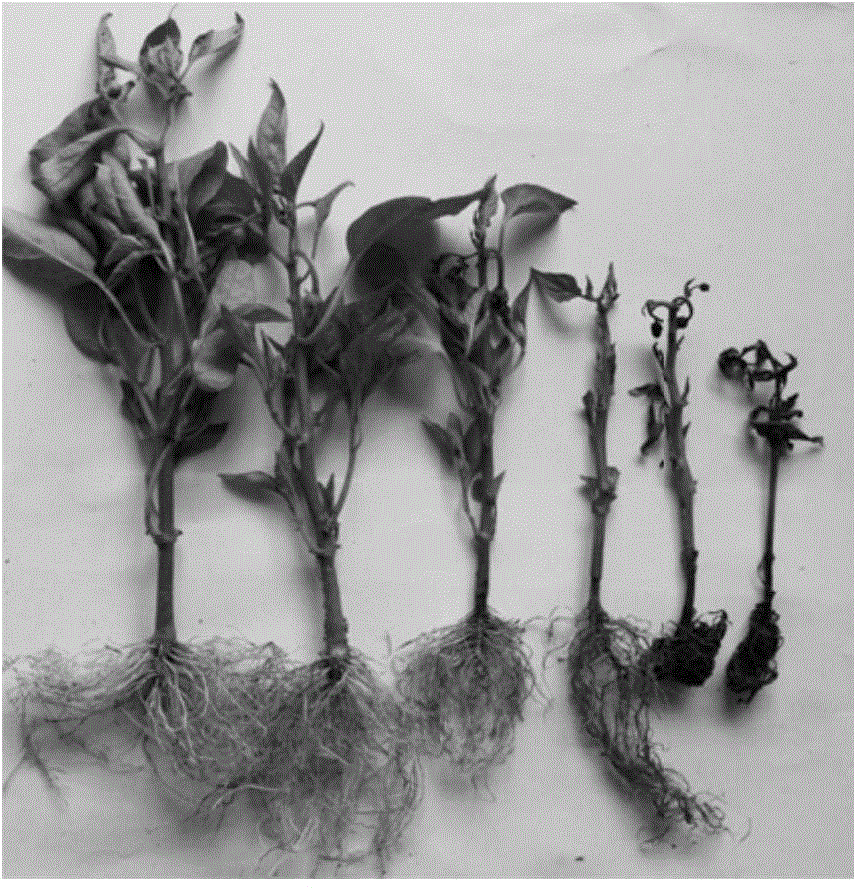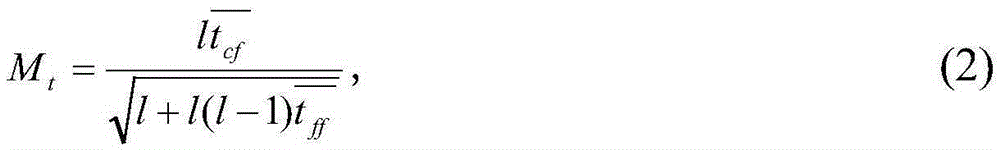Patents
Literature
149 results about "Gene pool" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
The gene pool is the set of all genes, or genetic information, in any population, usually of a particular species.
Aflatoxin nano antibody gene pool, construction method and application of aflatoxin nano antibody gene pool as well as aflatoxin B1 nano antibody 2014AFB-G15
ActiveCN103866401AEasy to buildResistant to organic reagentsImmunoglobulins against fungi/algae/lichensBiological testingGene poolAntibody variable region
The invention relates to an aflatoxin nano antibody gene pool, a construction method and application of the aflatoxin nano antibody gene pool as well as an aflatoxin B1 nano antibody 2014AFB-G15. The aflatoxin nano antibody gene pool is prepared by extracting RNA (ribonucleic acid) in alpaca blood after immunization of an aflatoxin B1 antigen, performing specific amplification on a variable region gene of an alpaca heavy chain antibody by adopting an RT-PCR (reverse transcription-polymerase chain reaction) method to obtain an aflatoxin nano antibody VHH gene, and then performing transformation after connecting with a pCANTAB5E (his) vector. The aflatoxin B1 nano antibody 2014AFB-G15 obtained by screening, disclosed by the invention, has the characteristics of organic reagent resistance, high temperature resistance and the like, and is good in stability; the IC50 (half maximal inhibitory concentration) of the aflatoxin B1 nano antibody 2014AFB-G15 to the aflatoxin B1 is 0.66ng / mL, and the cross reactivity of the aflatoxin B1 nano antibody 2014AFB-G15 to the aflatoxins B2, G1,G2 and M1 is 22.6%, 0.95%, 32.1% and 26% respectively.
Owner:INST OF OIL CROPS RES CHINESE ACAD OF AGRI SCI
Bovine germline D-genes and their application
ActiveUS7196185B2Large capacitySugar derivativesMicrobiological testing/measurementDiseaseImmunocompetence
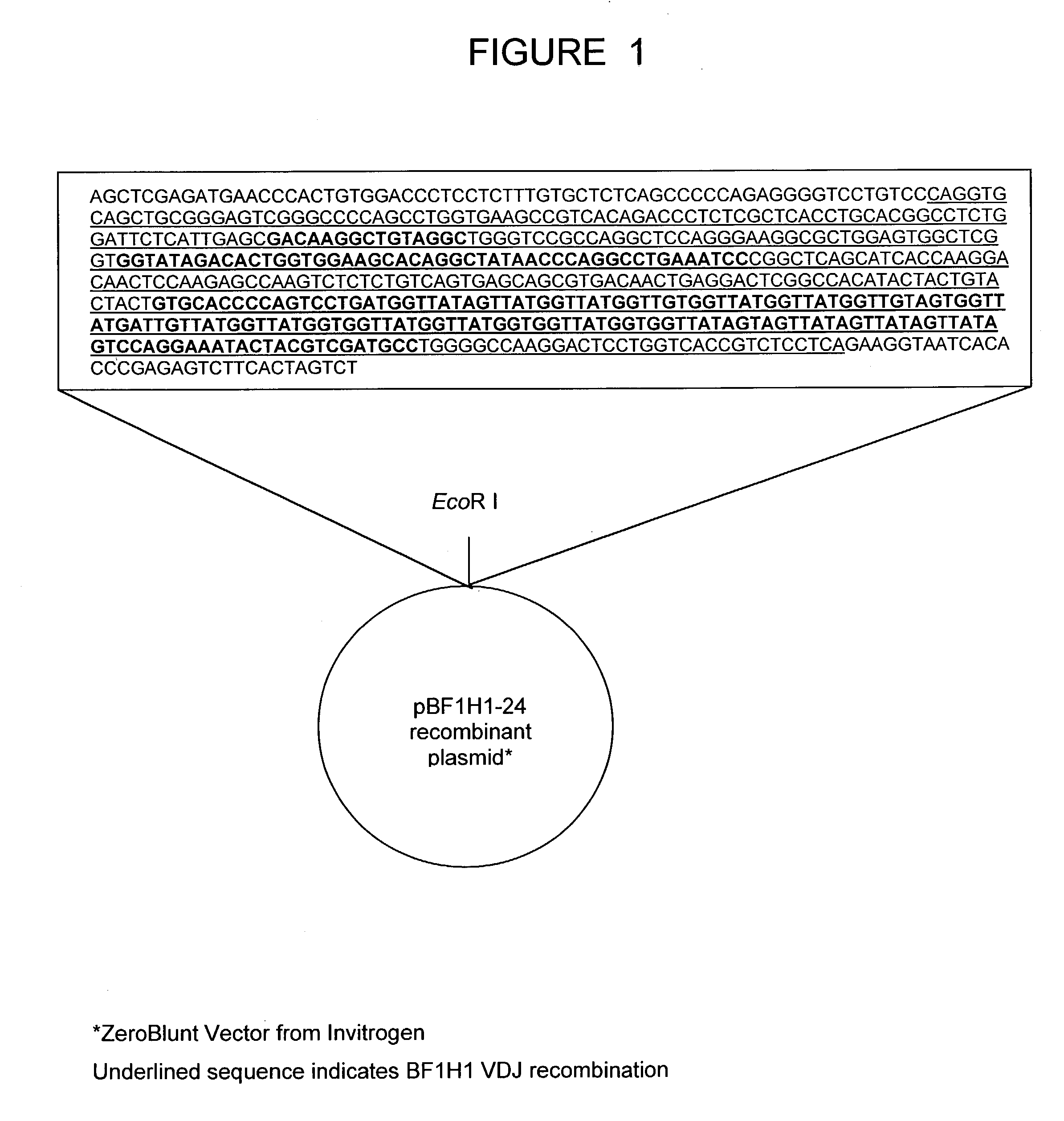
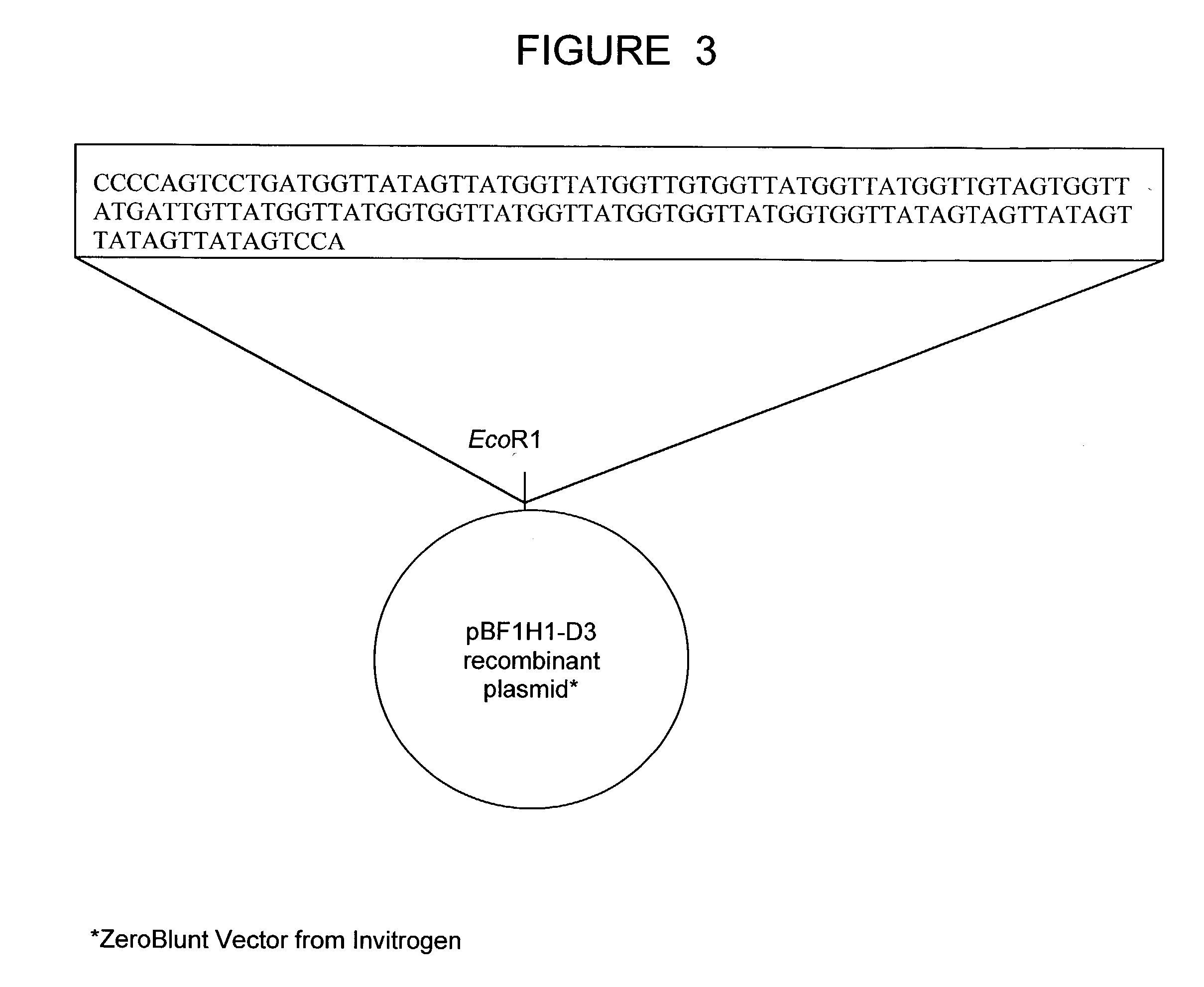
The present invention relates to a bovine VDJ cassette (BF1H1) that provides the novel ability to develop chimeric immunoglobulin molecule capable of incorporating both linear T cell epitope(s) (CDR1H and CDR2H) as well as conformational B cell epitope(s) (exceptionally long CDR3H). Further, multiple epitopes can be incorporated for development of multivalent vaccine by replacing at least a portion of an immunoglobulin molecule with the desired epitope such that functional ability of both epitope(s) and parent VDJ rearrangement is retained. The antigenized immunoglobulin incorporating both T and B epitopes of interest is especially useful for development of oral vaccines for use in humans apart from other species including cattle. The long CDR3H in BF1H1 VDJ rearrangement originates from long germline D-genes. The novel bovine germline D-genes provide unique molecular genetic marker for sustaining the D-gene pool in cattle essential for immunocompetence via selective breeding. D-gene specific DNA probe permits typing and selection of breeding cattle stock for maximum gemline D gene pool for better health and disease prevention. The bovine D-genes are unique to cattle and, therefore, provide sensitive and specific forensic analytical tool using molecular biology techniques to determine tissues suspected of bovine origin.
Owner:KAUSHIK AZAD KUMAR +2
Human autogenous siRNA sequence, its application and screening method
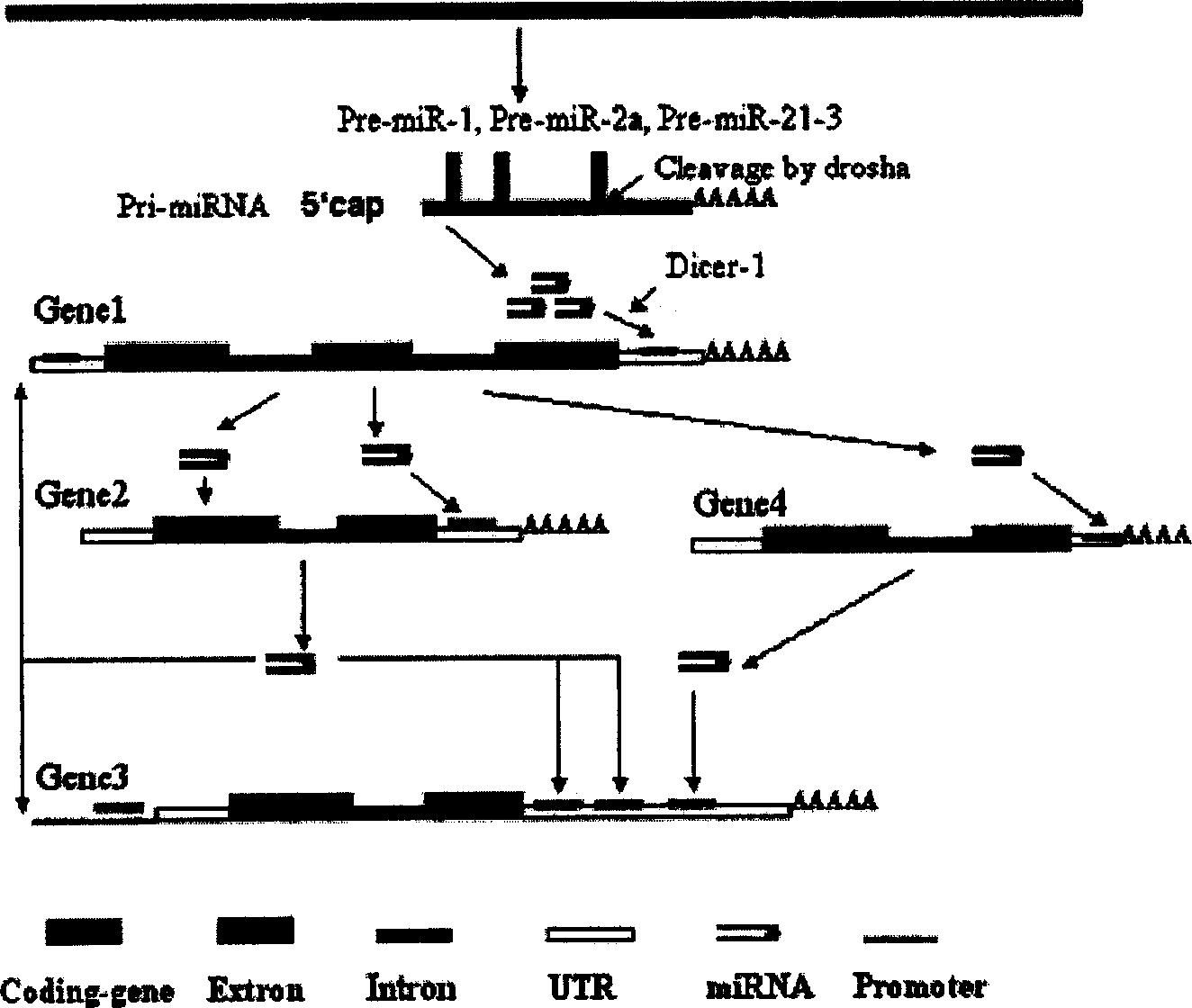
The invention relates to a computer algorithmic language and relative application software. It could rapidly select and predict internal source siRNA gene and siRNA molecule. Especially, it relates to an entire finding method for internal source siRNA gene and internal source siRNA molecule, and gaining 255 new siRNAs and target mRNAs from coding gene intron. The siRNA molecules could be used to research develop, differentiation and growth of cell, organization and organic, discuss the function and express adjusting network and discuss function of genes. It also could be used to develop medicines to prevent and cure kinds of disease.
Owner:INSITUTE OF BIOPHYSICS CHINESE ACADEMY OF SCIENCES
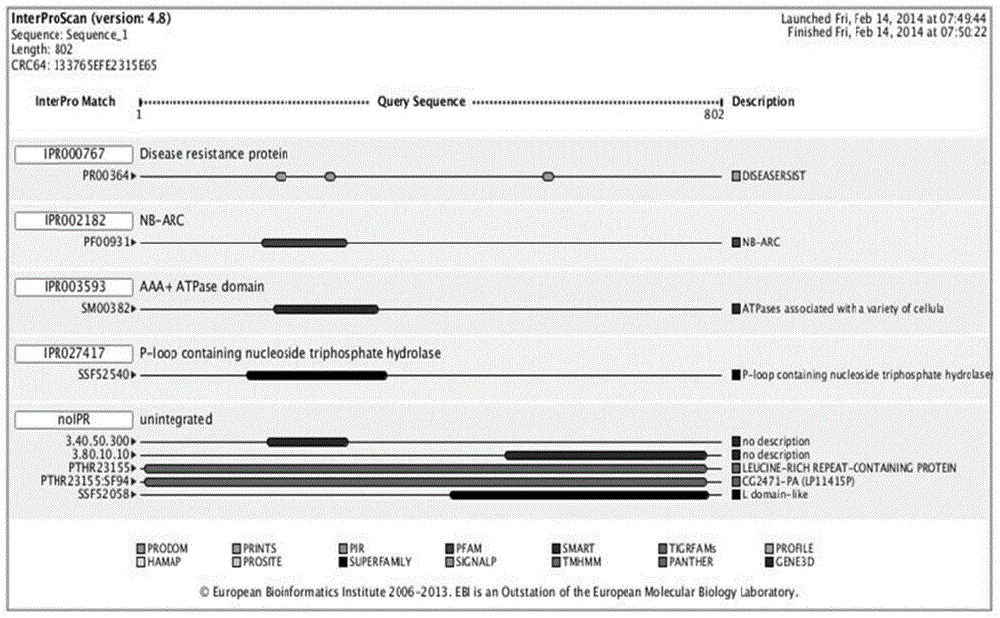
Method for obtaining capsicum phytophthora resistance candidate gene and molecular marker, and application
InactiveCN104560973AAccurate identificationLarge amount of data informationMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyData information
The invention relates to a method for obtaining a capsicum phytophthora resistance candidate gene and a molecular marker, and application. The method is used for obtaining the capsicum phytophthora resistance candidate gene by utilizing capsicum phytophthora transcriptome and whole-genome sequencing data information, differentially-expressed gene identification, bioinformatics analysis, molecular marker development and phytophthora inoculation identification and belongs to the technical field of capsicum biology. The method comprises the following steps: sequencing a phytophthora resistant and susceptible gene pool transcriptome obtained after phytophthora inoculation of an F2 population constructed by capsicum highly-resistant and highly-susceptible phytophthora materials, performing expression analysis and functional annotation on differential genes, extracting DNAs (Desoxvribose Nucleic Acid) of a capsicum phytophthora highly-resistant and highly-susceptible phytophthora material genome, performing primer design and PCR (Polymerase Chain Reaction) amplification, performing sequence difference analysis and SNP site identification, performing SNP specific primer design and validity verification, and performing other steps to efficiently obtain the capsicum phytophthora resistance candidate gene and the molecular marker. According to the method, the capsicum phytophthora resistance candidate gene can be accurately identified, and the effective molecular marker can be developed.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
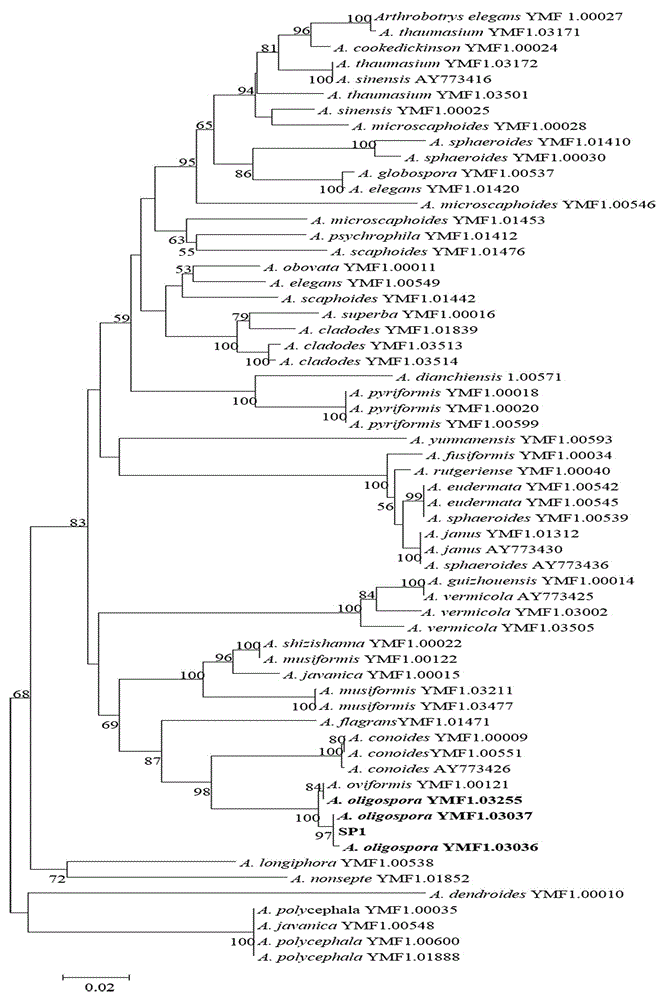
Method for identifying predator nematophagous hyphomycete arthrobotrys through DNA bar codes
InactiveCN105063761AEasy to expandEasy to compareLibrary creationSpecial data processing applicationsDNA barcodingGermplasm
The invention provides a method for identifying predator nematophagous hyphomycete arthrobotrys through DNA bar codes and belongs to the field of fungus species identification. According to the method, RPB2 genes serve as target DNA bar code genes for identifying the predator nematophagous hyphomycete arthrobotrys, combined arthrobotrys sample experiment data and target fungus data merged strategy is adopted to establish a high-cavity database, meanwhile, RPB2 genes to be identified are compared with a gene library, an identifying rule is established based on a system generation tree method of genetic distance method and clustering analysis of the Kimura-2-parameter probability to identify species. The method has the advantages that the RPB2 genes serve as the DNA bar codes most suitable for identifying the nematophagous hyphomycete arthrobotrys, and the method is universal and easy to amplify and compare. The identifying efficiency and the reliability and accuracy of the identifying method are greatly superior to those of a conventional DNA bar code method, and the method makes up for the blank of the nematophagous hyphomycete arthrobotrys DNA bar code molecular markers and the method provides a useful research tool for researches on germplasm resource excavation, biocontrol application and genetic diversity of nematophagous hyphomycete.
Owner:YUNNAN UNIV
Data mining technique with diversity promotion
ActiveUS8977581B1Digital computer detailsElectric digital data processingEvolutionary data miningCandidate Gene Association Study
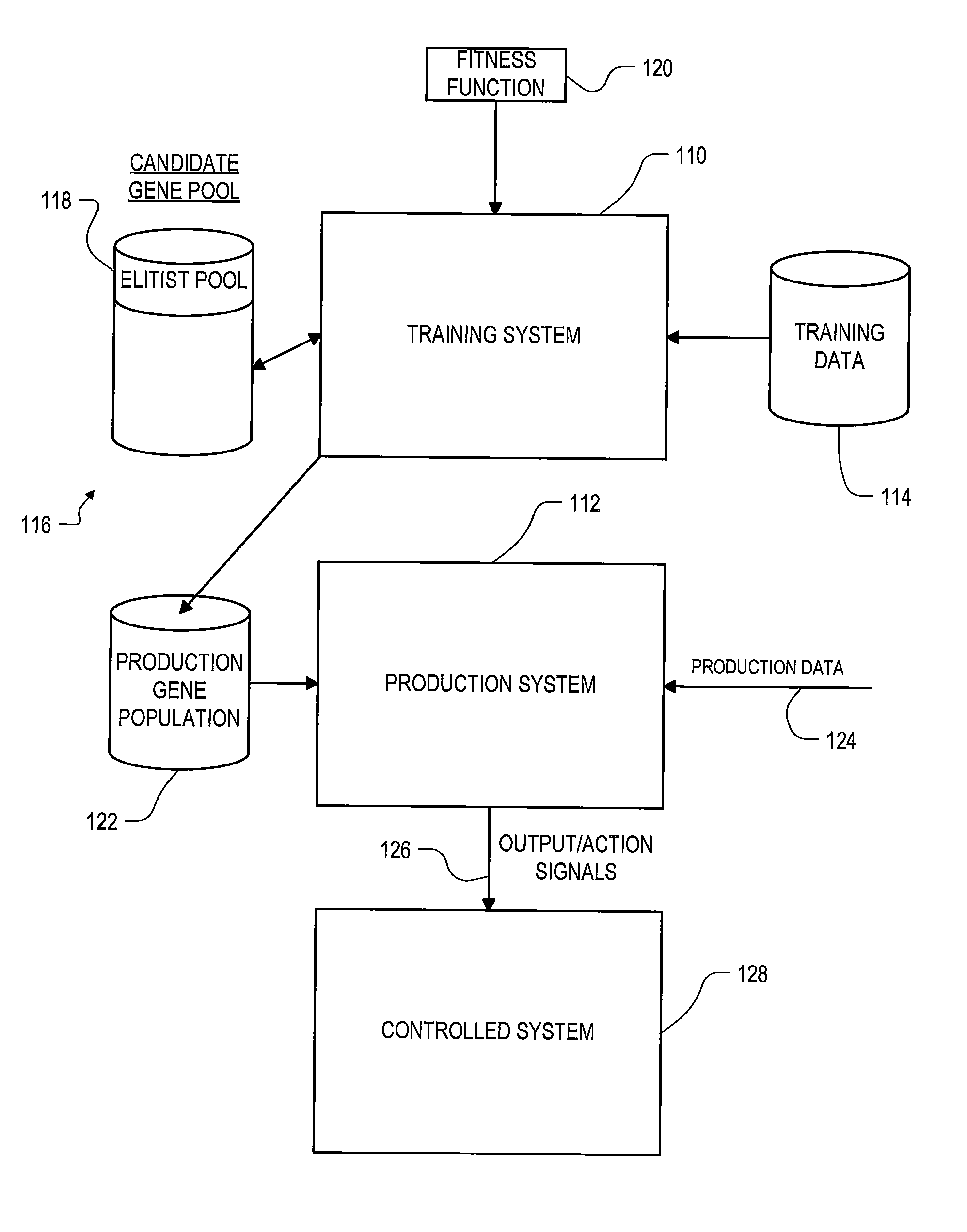
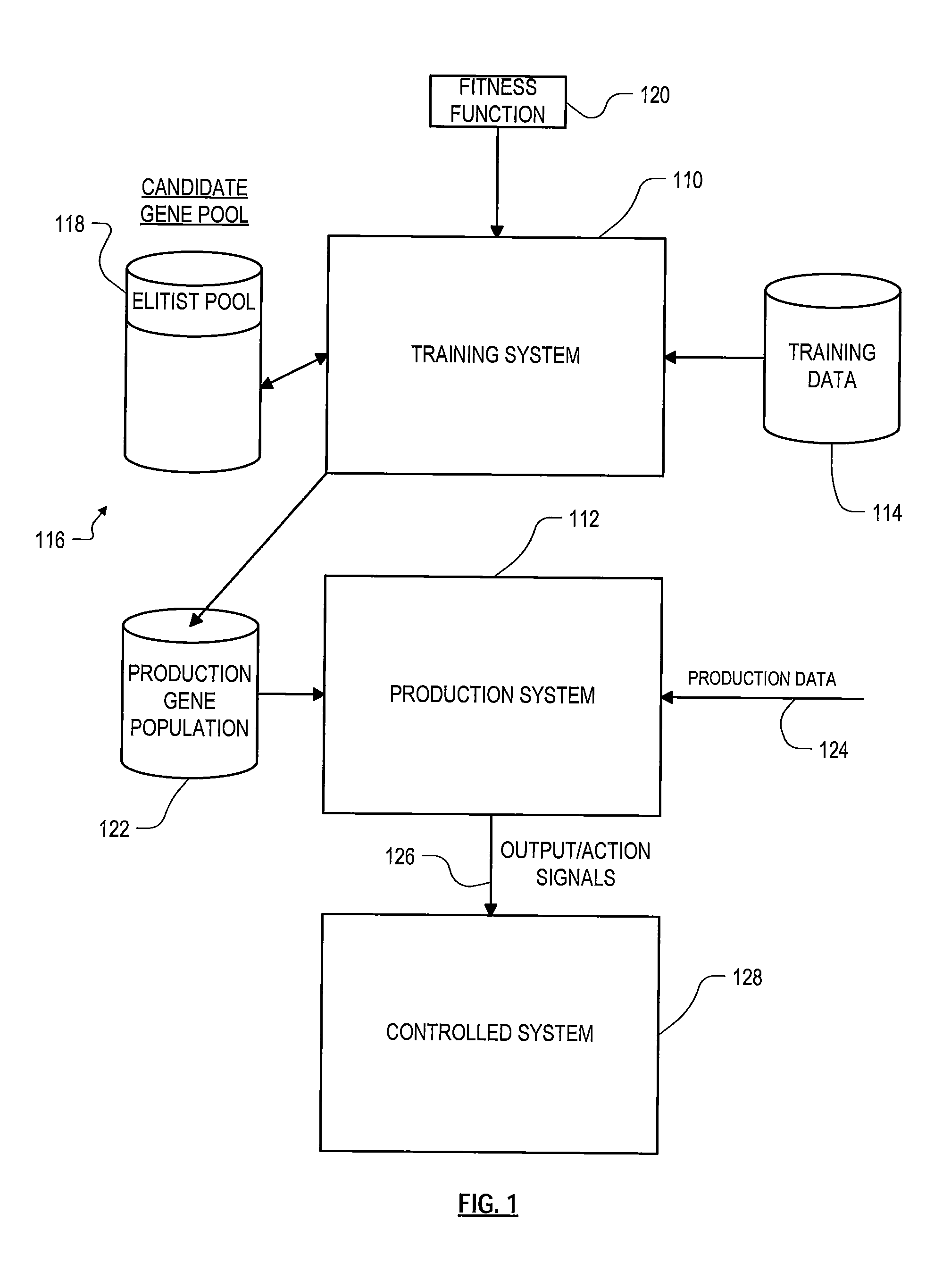
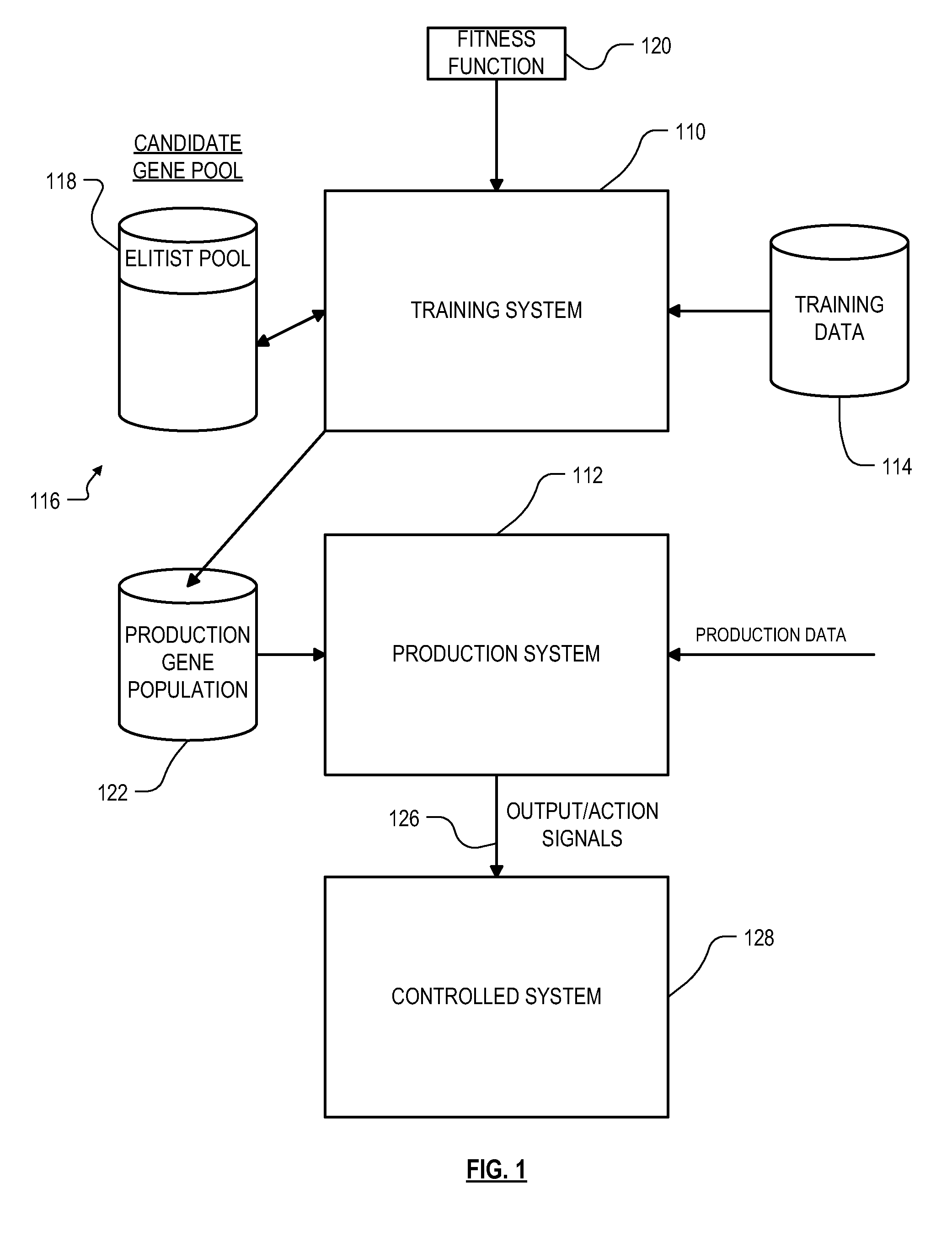
Roughly described, a computer-implemented evolutionary data mining system includes a memory storing a candidate gene database in which each candidate individual has a respective fitness estimate; a gene pool processor which tests individuals from the candidate gene pool on training data and updates the fitness estimate associated with the individuals in dependence upon the tests; and a gene harvesting module for deploying selected individuals from the gene pool, wherein the gene pool processor includes a competition module which selects individuals for discarding in dependence upon both their testing experience level and a diversity measure of individuals in the gene pool.
Owner:COGNIZANT TECH SOLUTIONS U S CORP
Method for screening anthracycline cardiotoxicity genes through mRNA expression profiles and competitive endogenesis RNA expression profiles jointly
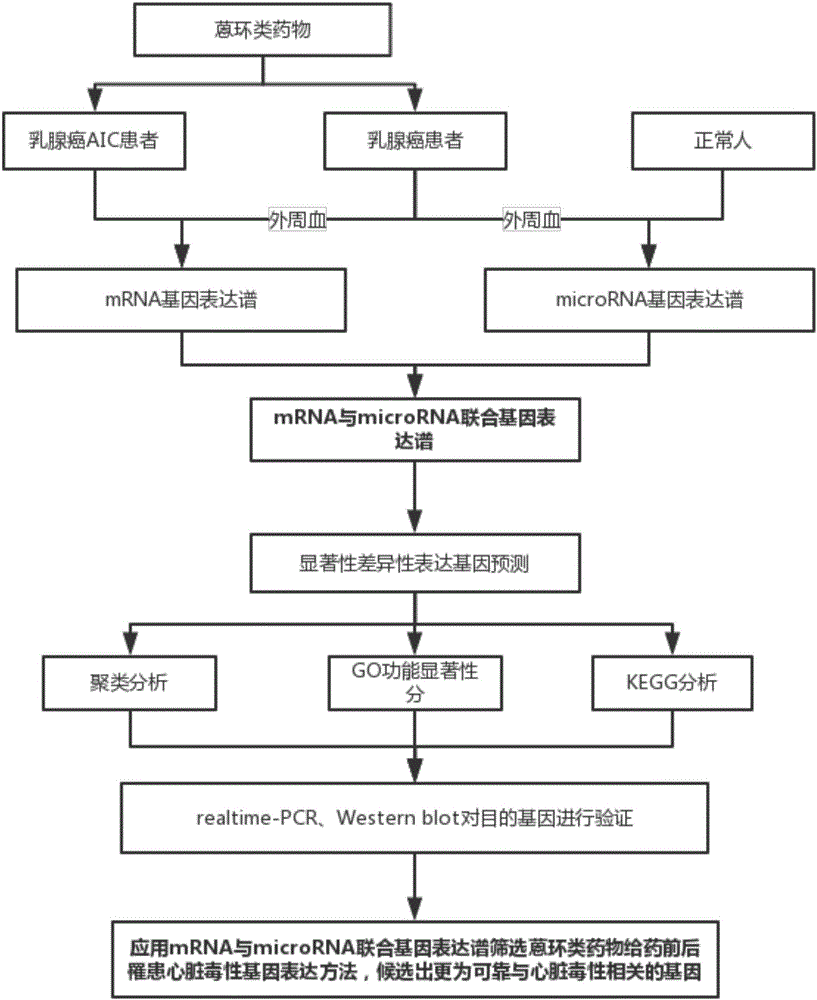
The invention relates to a method for screening anthracycline cardiotoxicity genes through mRNA expression profiles and competitive endogenesis RNA expression profiles jointly. The method includes the steps that expression values of a large quantity of genes are detected through mRNA and competitive endogenesis RNA expression profile chips, and the correlations of all the genes are ordered; the obtained mRNA feature genes and the obtained competitive endogenesis RNA feature genes are combined; a gene pool is subjected to further gene selection with a genetic algorithm; differential expression genes are selected by comparing the data between different samples of mRNA and competitive endogenesis RNA joint expression profiles of breast cancer patients, breast cancer AIC patients and able-bodied persons; the differential expression genes are predicted, and the genes with the scores larger than 140 and free energy smaller than 20 are selected as reliable target genes; genes with obvious differences are selected from the target genes, and constructed breast-cancer anthracycline cardiotoxicity attacking target genes are verified through the residual RNA. By means of the method, more reliable potential gene markers related to anthracycline cardiotoxicity can be selected as a candidate.
Owner:JINZHOU MEDICAL UNIV
Cucumber SNP marker and detection methods thereof
InactiveCN101591708AShorten development timeReduce verification timeMicrobiological testing/measurementDNA/RNA fragmentationF1 generationCucumber family
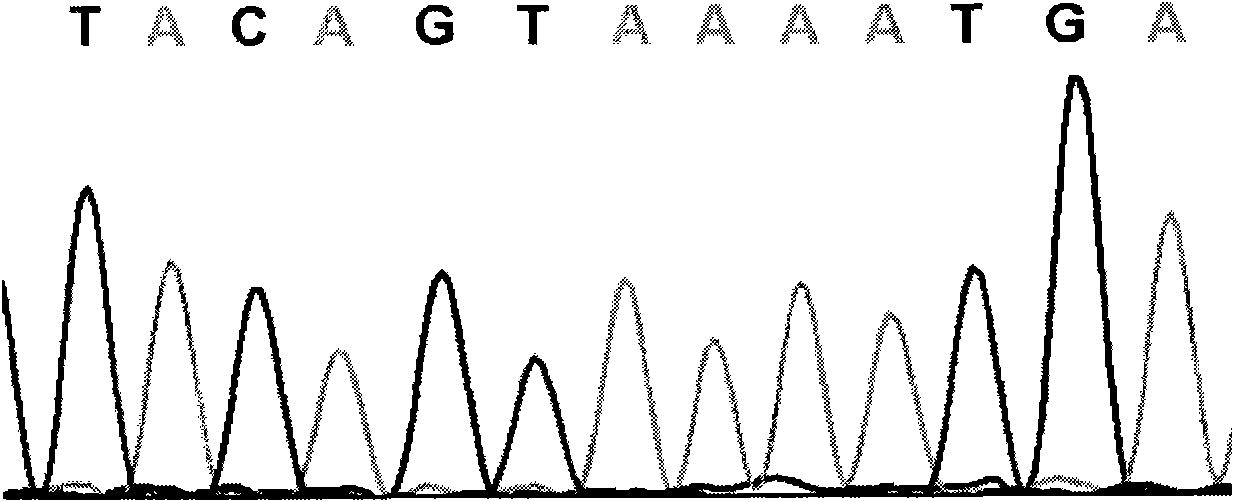
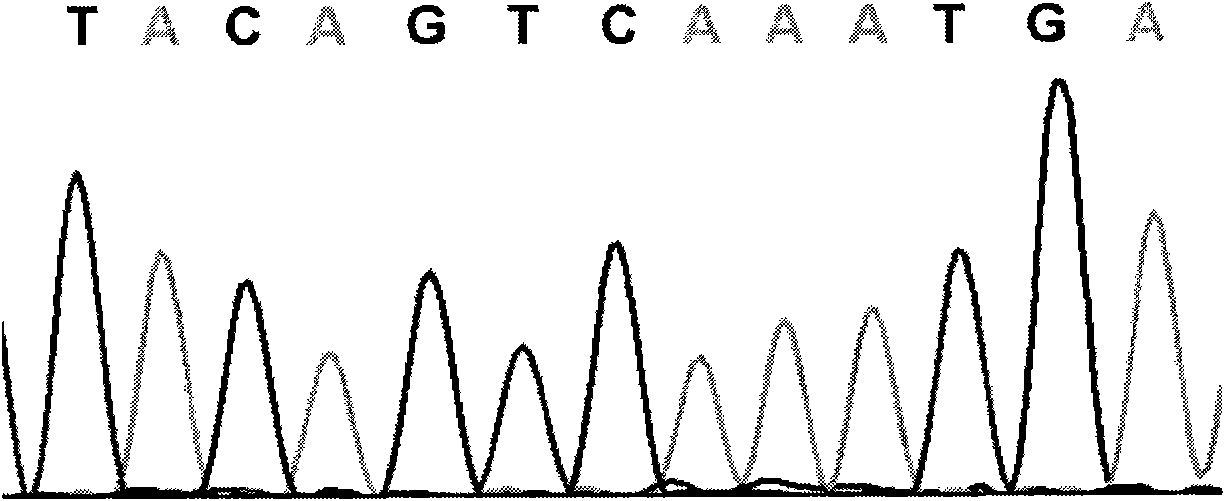
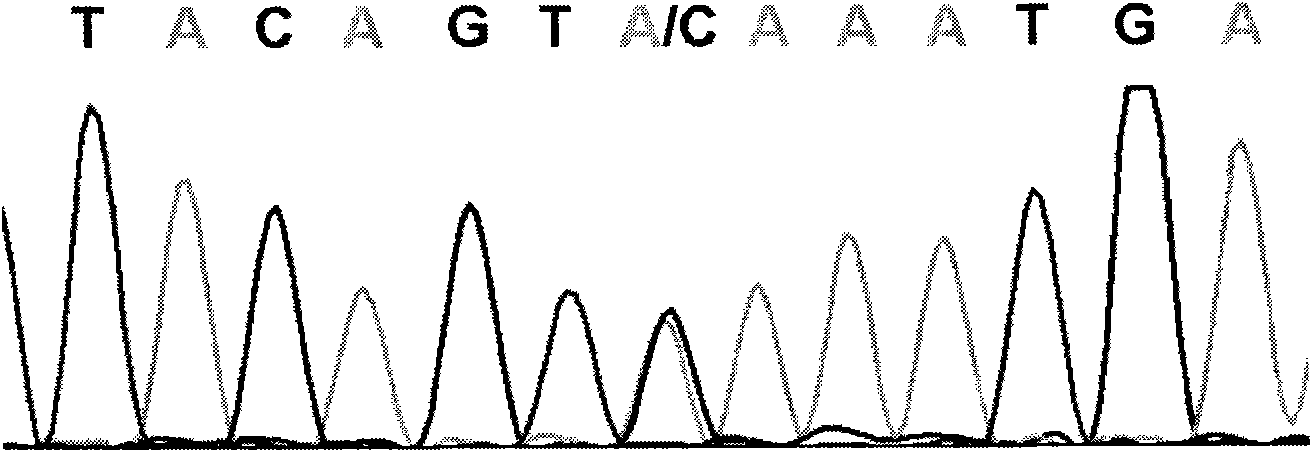
The invention relates to a cucumber SNP marker and detection methods thereof in the technical field of plant gene engineering. A method for identifying cucumber SNP locus comprises the following steps: selecting homozygous cucumber parents, and performing sequencing to determine candidate SNP locus; hybridizing to obtain F1 generation single plants, and performing sequencing; and detecting a peak shape chart of a F1 generation sequencing result to determine candidate locus with heterozygous peaks. A method for determining SNP marked polymorphic single plants in F2 generation separation group of cucumber comprises the following steps: selecting homozygous cucumber parents, and obtaining the F2 generation separation group; selecting recessive phenotype single plants in the F2 generation, mixing DNAs of the single plants in equal quantity, and construct a recessive gene pool; performing amplification and sequencing by an SNP primer, and detecting a peak shape chart of a sequencing result; and respectively performing amplification and sequencing on single plants in the recessive gene pool by the SNP primer, and detecting a peak shape chart of a sequencing result. The cucumber SNP marker has a sequence shown as SEQ ID NO:1. The invention shortens the development and testing time of the SNP marker, and reduces the detection cost of the marker.
Owner:SHANGHAI JIAO TONG UNIV
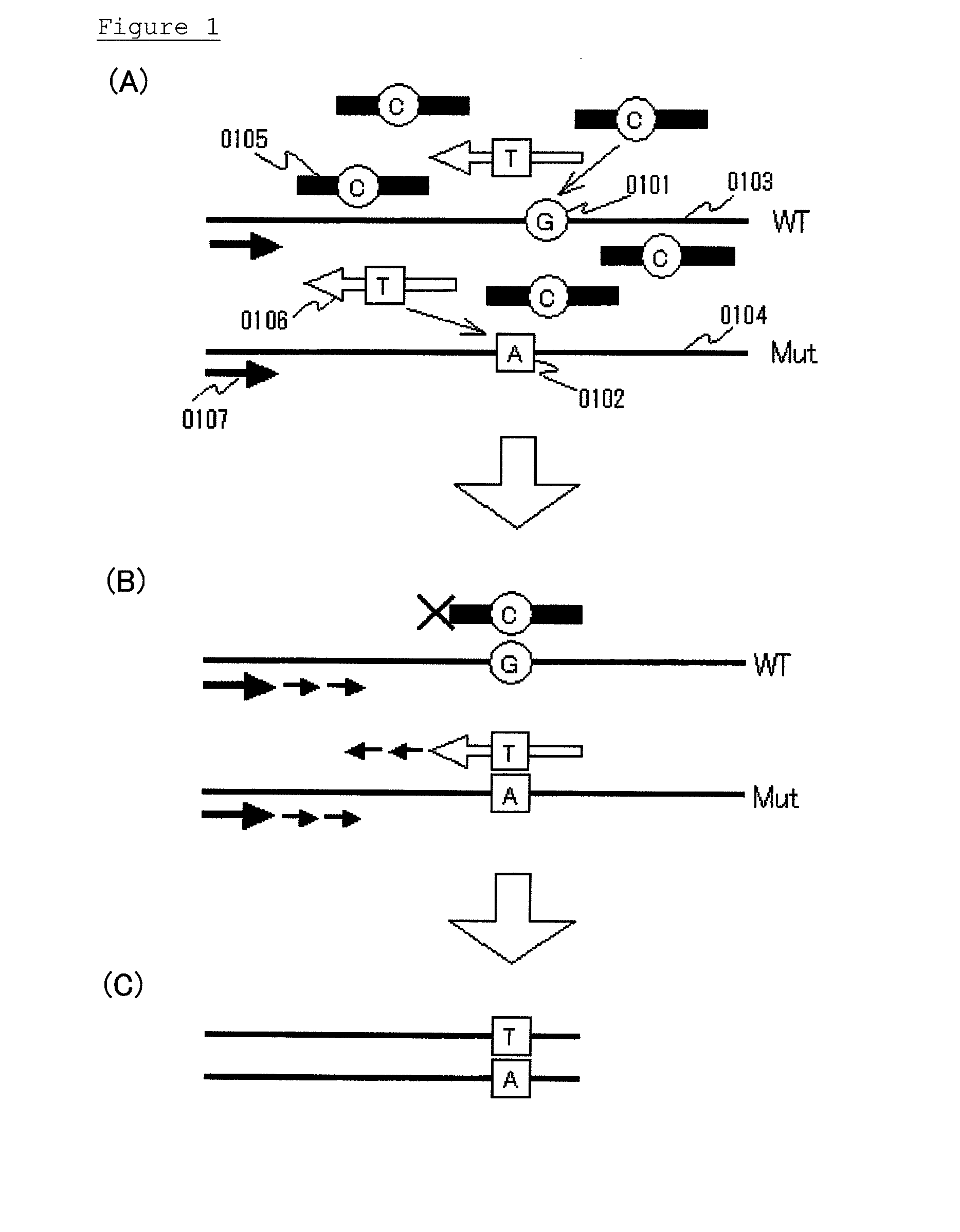
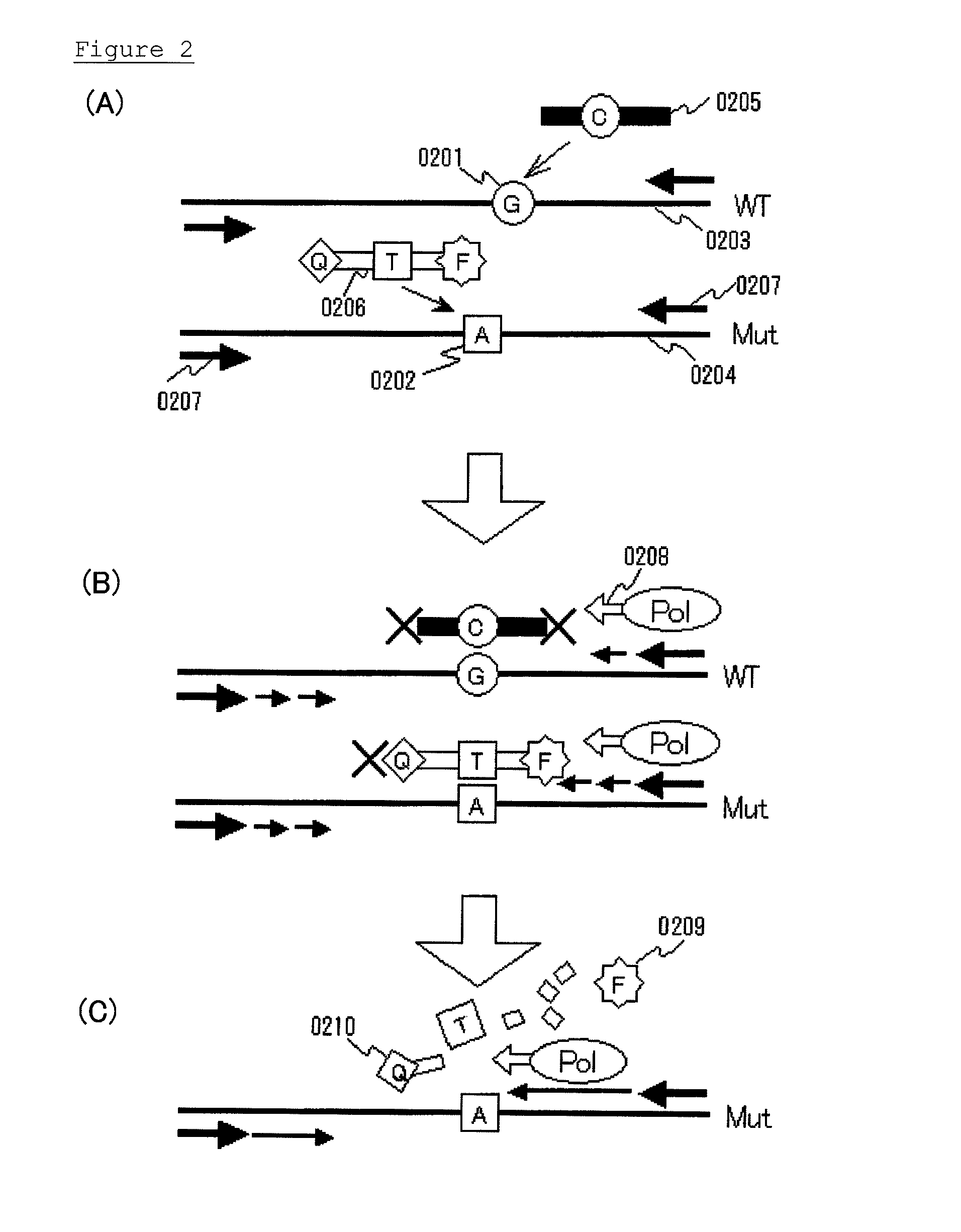
Highly sensitive method for detecting mutated gene
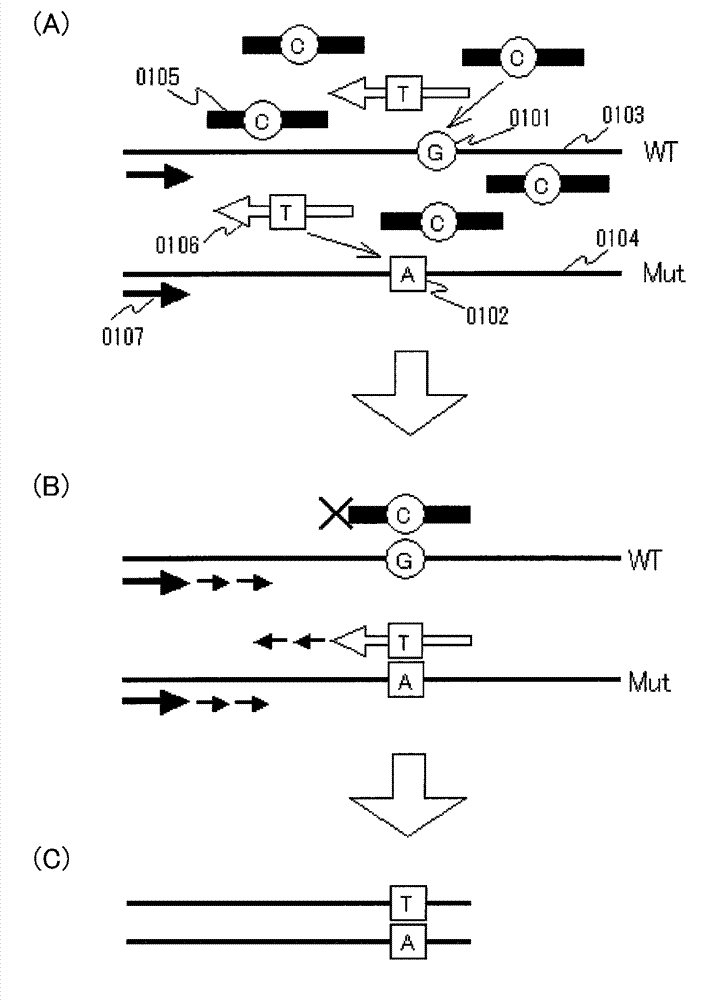
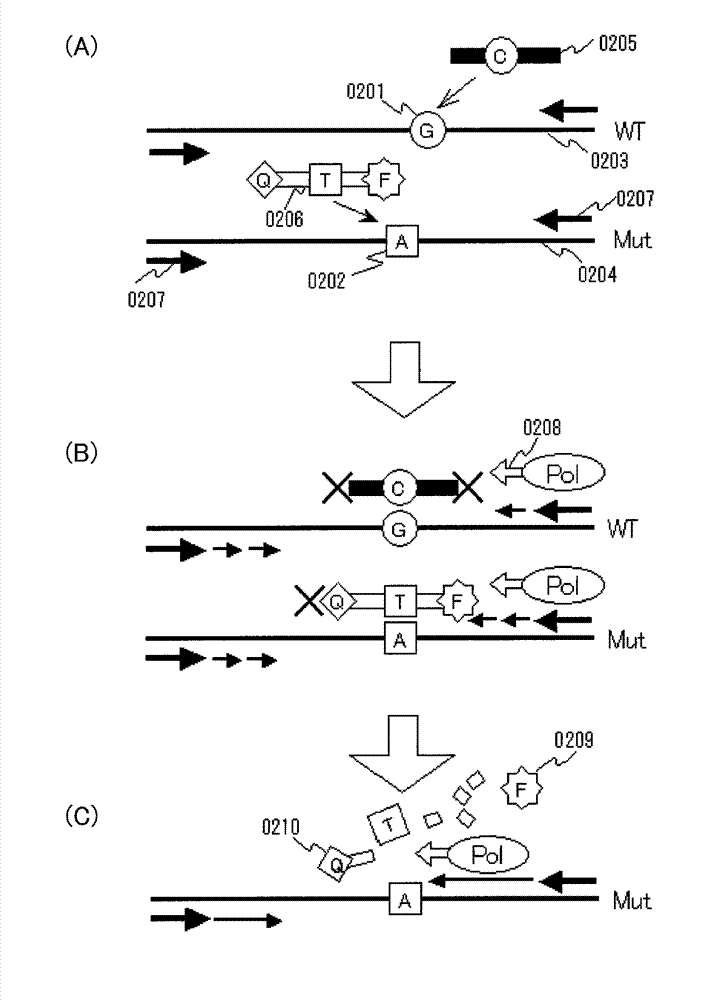
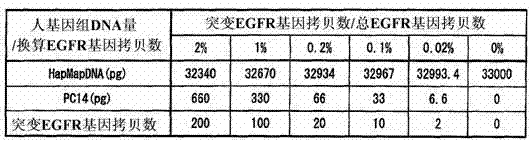
InactiveUS20130005589A1Improve accuracyHigh sensitivityMicrobiological testing/measurementLibrary screeningWild typeBiology
Various highly sensitive detection methods, particularly improved PNA-LNA-PCR clamp methods, are provided as methods for detecting the presence or absence of a mutated gene contained in a gene pool rapidly, in a simple manner, with high accuracy, and with high sensitivity. As a step before the main step for detection, a pre-amplification step comprising allowing (1) a clamp primer consisting of PNA which hybridizes with all or part of a target site having a sequence of a wild-type gene or a sequence complementary to the wild-type gene, (2) a primer capable of amplifying a region comprising a target site having a sequence of the mutated gene, and (3) the gene pool to coexist in a reaction solution for gene amplification, and selectively amplifying the region comprising a target site of the mutated gene by a gene amplification method.
Owner:MITSUBISHI CHEM MEDIENCE
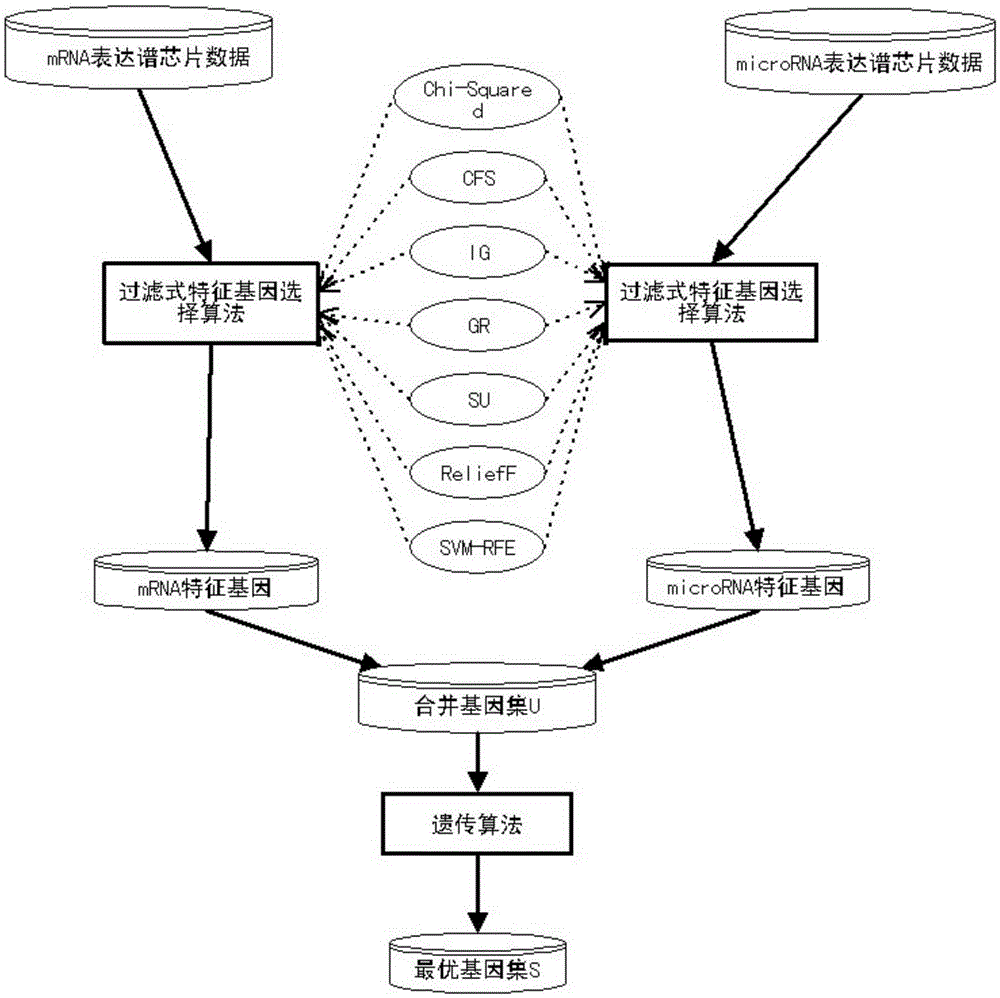
Tumor feature gene selection method combining mRNA and microRNA expression profile chips
InactiveCN105243296AImprove classification accuracyEasy to analyzeSpecial data processing applicationsGene selectionFiltration
The invention discloses a tumor feature gene selection method combining mRNA and microRNA expression profile chips. The method is implemented by the following steps of step 1, by using the mRNA and microRNA expression profile chips, detecting expression values of a large amount of genes, by adopting a filtration feature gene selection method, ranking relevance of all the genes, removing a large amount of low-relevance genes, and leaving a small amount of genes that are closely related to tumor classification; step 2, combining mRNA and microRNA feature genes obtained by adopting the filtration feature gene selection method to form a gene pool U; and step 3, by adopting a genetic algorithm, further selecting genes for the gene pool, eliminating redundant genes, and performing a search to obtain an optimal gene set S with optimal features. The method is simple in step and high in result accuracy.
Owner:LISHUI UNIV
Detection method of bot program
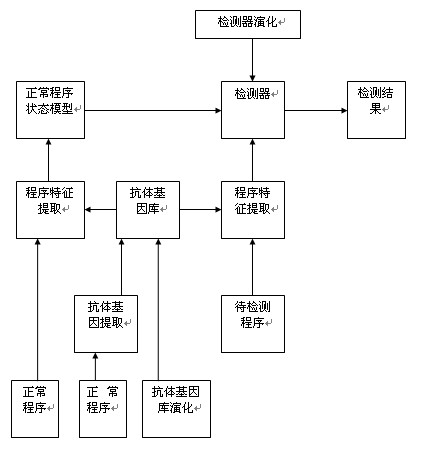
ActiveCN101930517ACan't solveSolve the outstanding contradiction of polymorphic bot synchronizationPlatform integrity maintainanceFeature extractionState model
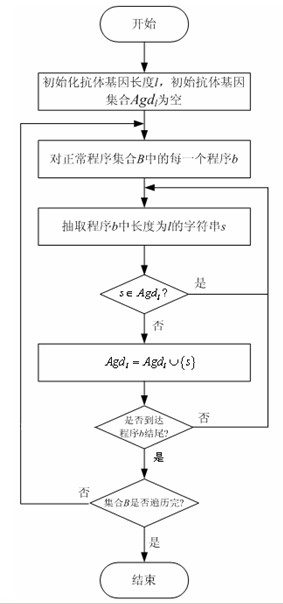
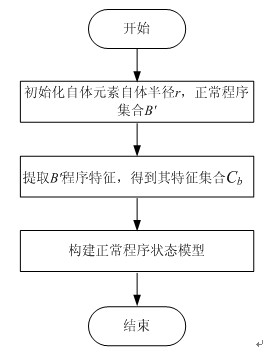
The invention discloses a detection method of a bot program and belongs to the technical field of information safety, and the method comprises the following steps: extracting antibody genes of a normal program set B, constructing antibody gene sets Agdl, and forming an antibody gene library Agd by the antibody gene sets Agdl of different antibody gene lengths; carrying out feature extraction on a normal program set B' by the antibody gene sets Agdl and constructing a normal program state model; generating detectors by normal program state sets Cb and generating a detector set by the detectors; detecting the bot program by the detector set; and evolving the antibody gene library and the detectors dynamically. The method can not only identify known bot programs but also discover new bot programs or variations of the known bot programs through self-learning and evolvement in a computer environment which changes in real time, thus effectively solving the key issue that a feature code library of computer viruses can not be synchronous with the multistate bot programs.
Owner:四川通信科研规划设计有限责任公司
QTL-seq based method for mining cold-tolerant gene of Dongxiang wild rice
The invention discloses a QTL-seq based method for mining a cold-tolerant gene of Dongxiang wild rice. According to the method, cultivated rice 'Xieqing Zao B' is taken as a receptor parent, namely, a female parent; the Dongxiang wild rice is taken as a male parent; a recombinant inbred line BC1F10 obtained by distant hybridization and continuous backcrossing is subjected to cold tolerance identification; obtained 20 cold-tolerant strains form a cold-tolerant gene pool; and 20 cold-sensitive strains form a cold-sensitive gene pool. The two gene pools are subjected to high-throughput sequencing and QTL-seq analysis to obtain differential SNP makers and cold-tolerant character related QTL loci of the two gene pools; candidate cold-tolerant genes are subjected to fluorescent quantitative PCR detection; and finally the Dongxiang wild rice is detected to contain five cold-tolerant related genes by identification. According to the method, the cold-tolerant genes of the Dongxiang wild rice can be quickly and accurately detected and mined, a powerful evidence can be provided for comprehensively illuminating a genetic mechanism of cold tolerance character of the Dongxiang wild rice, a foundation is laid for cloning and functional analysis of the cold-tolerant genes of the Dongxiang wild rice, and the breeding practice of the cold-tolerant genes of the Dongxiang wild rice is facilitated.
Owner:JIANGXI NORMAL UNIV
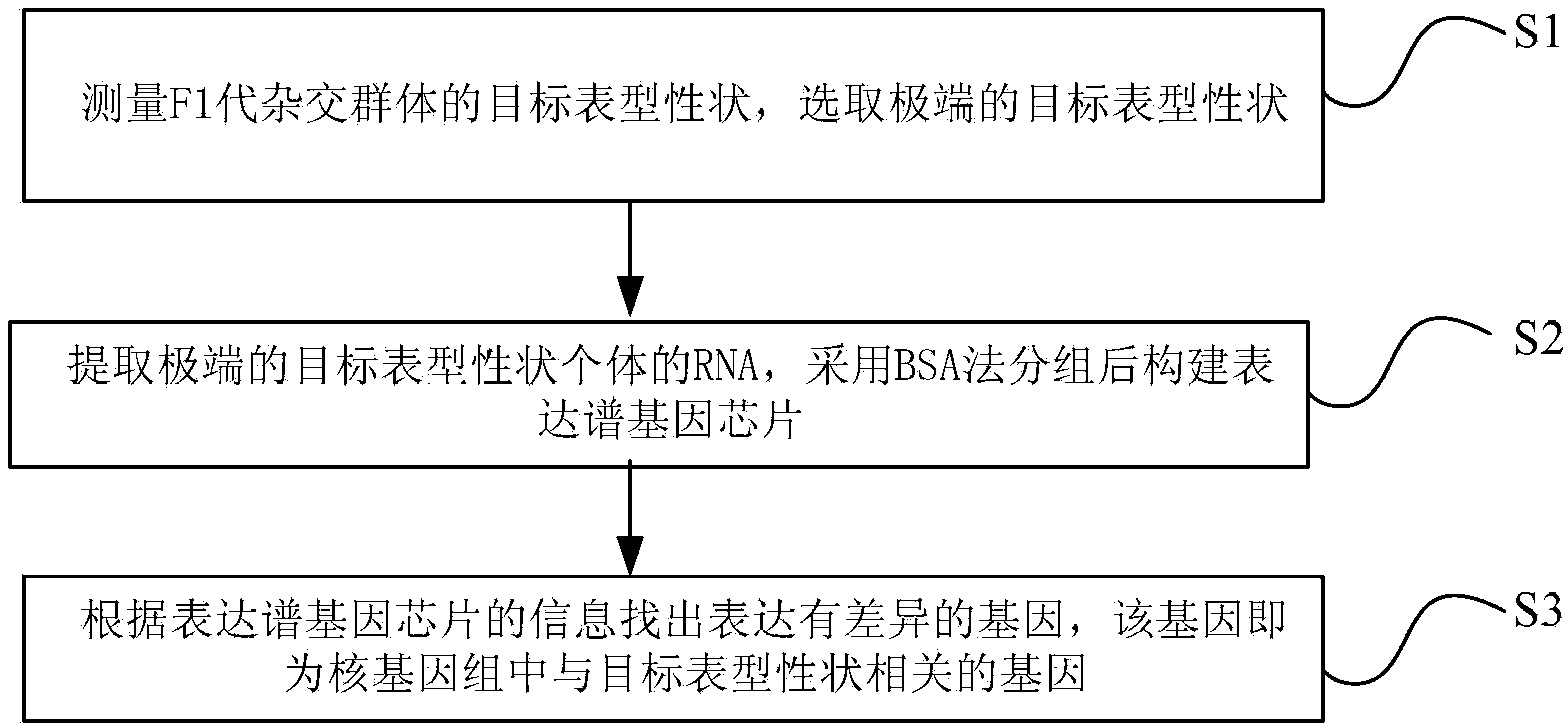
Method of detecting phenotypic character related genes in nuclear genome
The invention discloses a method of detecting phenotypic character genes related in a nuclear genome. The method includes: S1) measuring a target phenotypic character of an F1-generation hybrid population and selecting an extreme target phenotypic character; S2) extracting RNA of the individuals with the extreme target phenotypic character, grouping by a BSA method, and constructing an expression gene chip; and S3) finding differentially expressed genes according to information of the expression gene chip, wherein the genes are the phenotypic character related genes in the nuclear genome. By application of the technical scheme, the target phenotypic character of the F1-generation hybrid population is screened by a plurality of times of measurement, the individuals with the stable extreme target phenotypic character are selected, mixing and grouping are performed by adoption of the BSA method and gene pools are constructed, thus highlighting differences of the target phenotypic character. A large number of genes are hybridized at the same time by combination of a gene chip technology. The differentially expressed genes related to the target phenotypic character are screened through analyzing chip results.
Owner:BEIJING FORESTRY UNIVERSITY
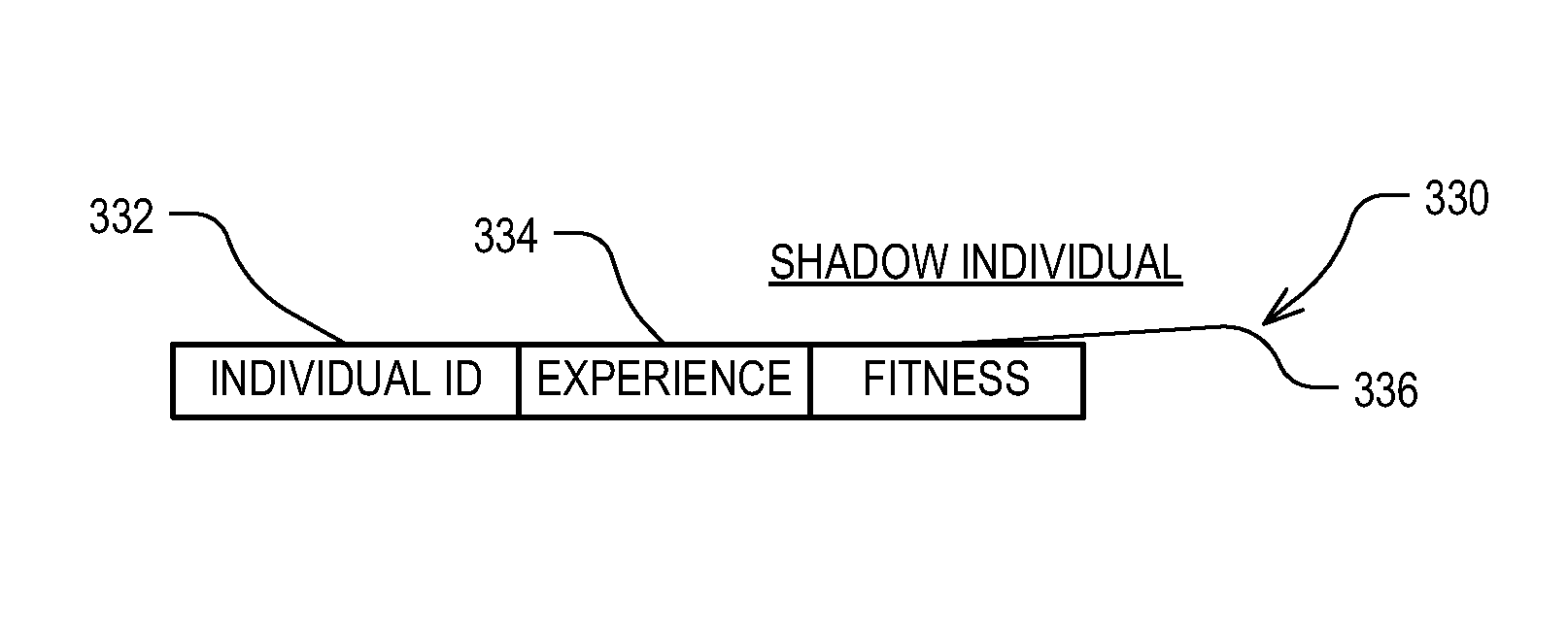
Data mining technique with shadow individuals
ActiveUS9256837B1Waste of scarceWell formedMachine learningSpecial data processing applicationsEvolutionary data miningCandidate Gene Association Study
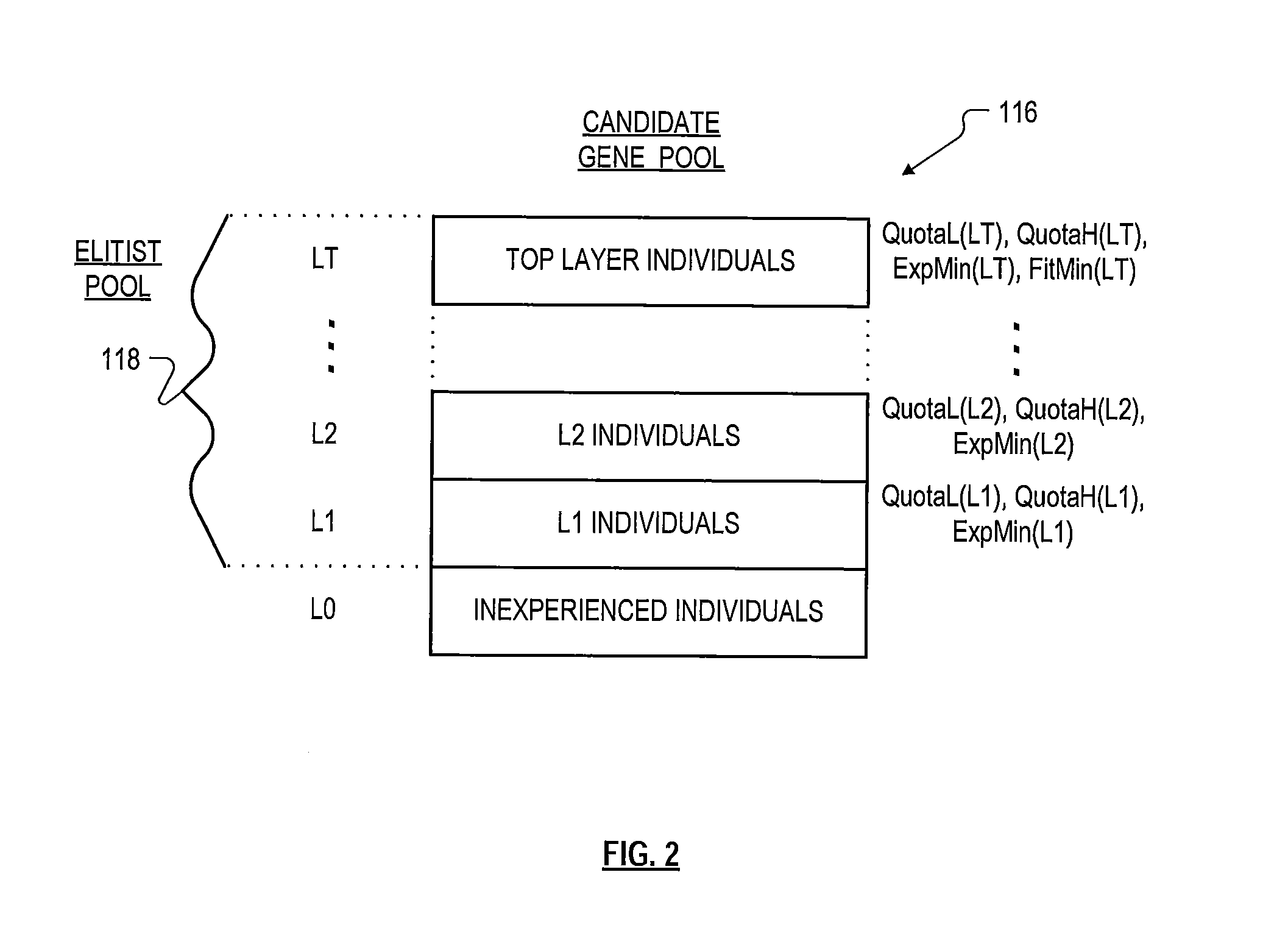
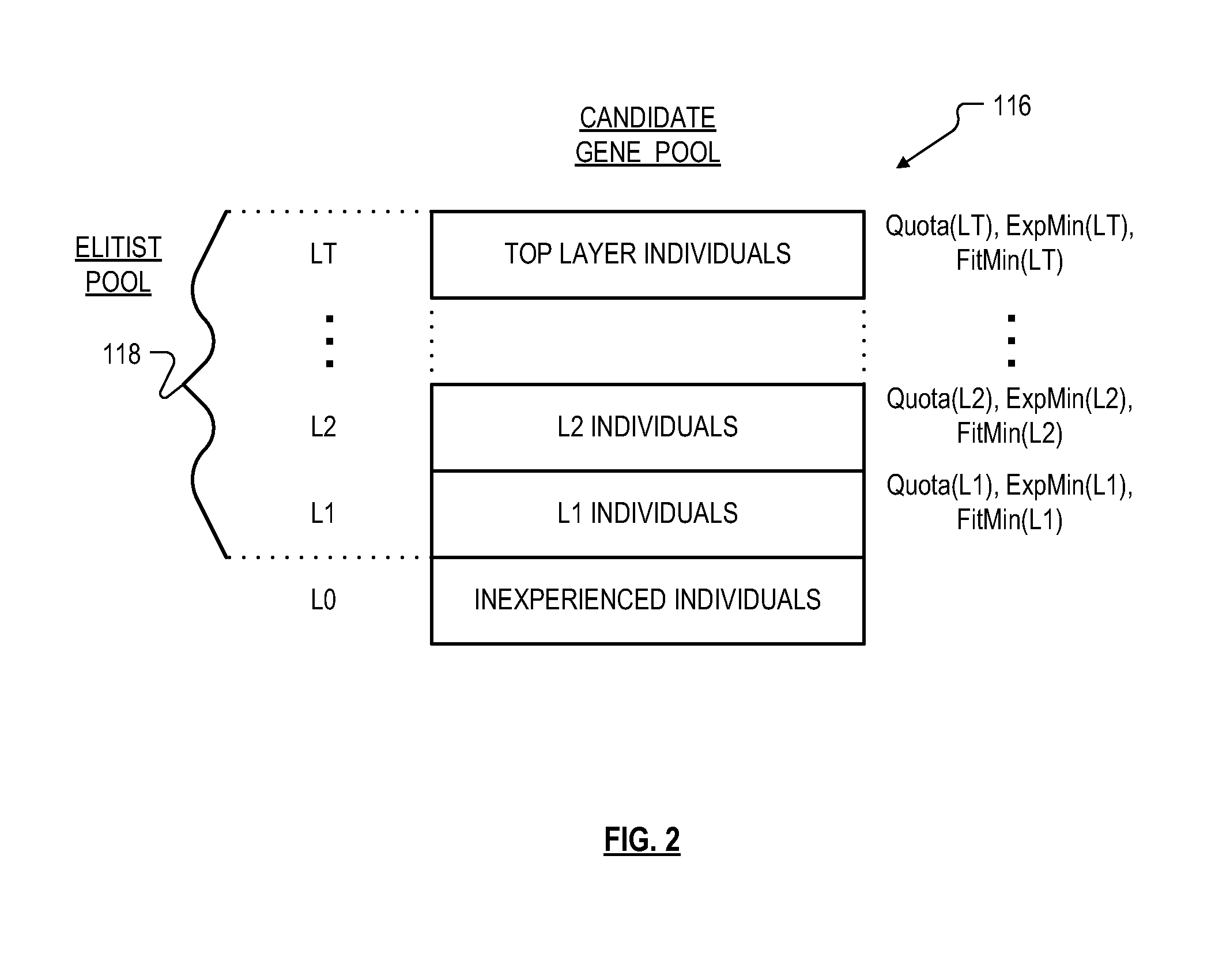
Roughly described, a computer-implemented evolutionary data mining system includes a memory storing a candidate gene database containing active and shadow individuals; a gene pool processor which tests only active individuals on training data and updates their fitness estimates; a competition module which selects individuals (both active and shadow) for discarding from the gene pool in dependence upon both their updated fitness estimate and their testing experience level; and a gene harvesting module providing for deployment selected ones of the individuals from the gene pool. The gene database has an experience layered elitist pool, and individuals to compete only with other individuals in their same layer. Shadow individuals are created in each layer for active individuals that survive all competition with the layer before their testing experience exceeds the testing experience range for the layer.
Owner:COGNIZANT TECH SOLUTIONS U S CORP
DNA technology based encryption method
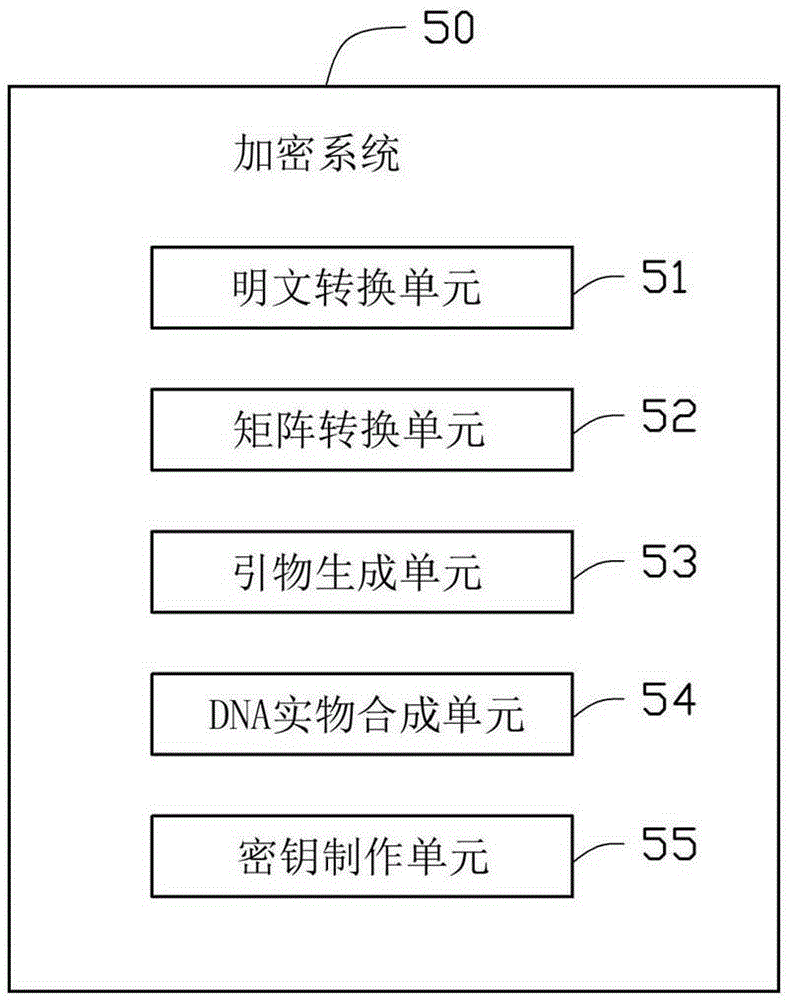
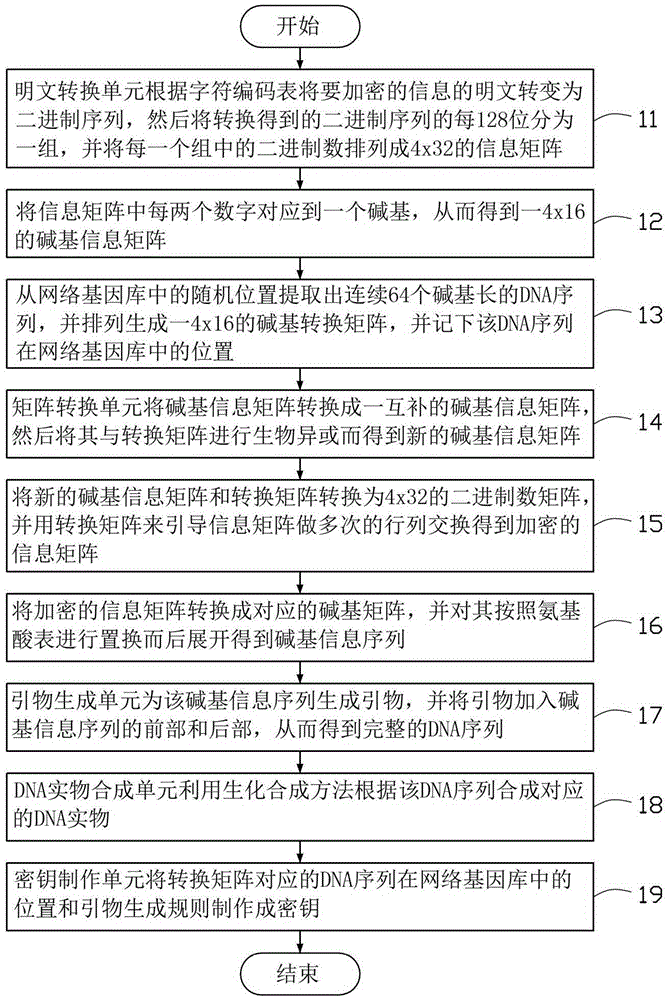
InactiveCN106817218AKey distribution for secure communicationEncryption apparatus with shift registers/memoriesPlaintextComputer science
A DNA technology based encryption method includes steps of converting plaintext of a to-be-encrypted message into binary sequences and dividing the binary sequences into a plurality of groups and arranging each group of the sequences into an information matrix; converting each two bits of the matrix into a corresponding basic group so as to converting the information matrix into a basic group information matrix; extracting a DNA sequence from a random position in a network gene pool and arranging the DNA sequence into a basic group converting matrix, performing conversion on the basic group information matrix by using the basic group converting matrix so as to obtain an encrypted basic group information matrix, unfolding the encrypted basic group information matrix so as to obtain a basic group information sequence; generating a primer for the basic group information sequence and adding the primer to the front part and the rear part of the basic group information sequence so as to obtain a complete DNA sequence; and synthesizing a corresponding DNA entity according to the DNA sequence by utilizing a biochemical synthesizing method.
Owner:AMBIT MICROSYSTEMS (SHANGHAI) LTD
Breeding method by utilizing distant hybridization and method for constructing gene library
PendingCN109197571ARich genetic baseFast aggregationPlant genotype modificationF1 generationGenomic library
The invention discloses a breeding method by utilizing distant hybridization and a method for constructing a gene library. The breeding method includes: 1) distant hybridization: performing interspecific distant hybridization on upland cotton, sea island cotton, wild cotton and Gossypium arboretum to obtain distant hybridization generations; 2) quick purification of the distant hybridization generations: with a hemigamy material being a female parent and the distant hybridization generations as a male parent, performing chromosome doubling to a haplobiont in the F1 generation for homozygosis;3) determining a core parent; 4) introduction, expression and screening of insecticidal gene: hybridizing the core parent and an insecticidal species to obtain F2 and BC1 generations, planting the generations, and performing screening by combination of field insect survey and kanamycin detection; 5) selective breeding of cottons in different species: according to the insecticidal character, disease resistant character, yielding ability, prematurity and fiber quality, comprehensively judging the species and classifying the species. The method can accelerate stabilization of the target characters and acquire the species with the target characters, and can be used for constructing the breeding gene library without different target characters and creating conditions for the selective breeding.
Owner:COTTON RES INST HEBEI ACAD OF AGRI & FOREST SCI
Highly sensitive mutated gene detection method
InactiveCN102959091AMicrobiological testing/measurementRecombinant DNA-technologyRegioselectivityWild type
Disclosed are various highly sensitive detection methods, particularly improved PNA-LNA-PCR clamp methods, as methods for detecting the presence or absence of a mutated gene included in a gene pool rapidly, in a simple manner, with high accuracy, and with high sensitivity. As a previous step for the main detection step, a previous amplification step is carried out, wherein the previous amplification step comprises allowing (1) a clamp primer which comprises a PNA that can hybridize with the entire region or a part of a target site comprising the sequence for wild-type gene or a sequence complementary to the wild-type gene, (2) a primer which can amplify a region containing a target site comprising the sequence for the mutated gene, and (3); the gene pool to coexist in a reaction solution for a gene amplification reaction and selectively amplifying the region containing the target site of the mutated gene by a gene amplification method.
Owner:MITSUBISHI CHEM MEDIENCE
Photoperiod sensitive type sudangrass hybridization breeding method
ActiveCN106856966AExtend the period of useImprove feed qualityData processing applicationsPlant cultivationBiotechnologyGermplasm
The invention discloses a photoperiod sensitive type sudangrass hybridization breeding method, and belongs to the technical field of forage variety breeding methods. Materials with parents or at least the female parent with photoperiod sensitive characteristics are selected for performing hybridization breeding, wherein according to F0, parent heading and blooming are induced in winter in Sanya, Hainan with 12 h or shorter day length, and hybridization grouping is completed; according to F1, authentication selection conditions are the same as that of F0; according to F2, photoperiod sensitive single plants are selected on the spring sowing condition in the North, and transplanted to Sanya, Hainan by adopting a rhizome propagation method for regrowth; regeneration stems of the plants of F2 are used for inducing blooming and seeding on the short-day condition in Sanya, Hainan, and the photoperiod sensitive characteristic verification is achieved by observing the time of a heading period; according to F3 to F5, repeated selection continues to be performed by means of the sunshine conditions of North-Sanya, Hainan; according to F6, seed reproduction is stabilized, and generations F7 and F8 are evaluated; the photoperiod sensitive sudangrass novel germplasm can be obtained. The novel germplasm bred through the breeding method enriches a sudangrass germplasm resource gene pool.
Owner:DRY LAND FARMING INST OF HEBEI ACAD OF AGRI & FORESTRY SCI
Primers, probes and method for detecting Mycobacterium tuberculosis drug-resistant gene mutation sites
ActiveCN105603084AStrong specificityHigh sensitivityMicrobiological testing/measurementMicroorganism based processesPectobacteriumGene mutation
The invention relates to primers, probes and a method for liquid-phase chip detection of Mycobacterium tuberculosis drug-resistant gene mutation sites. The primers, probes and method are used for detecting drug-resistant gene mutation sites in Mycobacterium tuberculosis for drugs isoniazide, rifampicin and fonoquantel. The isoniazide drug-resistant mutation sites are positioned in katG gene and inhA gene; the rifampicin drug-resistant mutation sites are positioned in rpoB gene; and the fonoquantel drug-resistant mutation sites are positioned in gyrA gene. The method comprises the following steps: respectively carrying out homology analysis according to the nucleotide sequences of the four drug-resistance related genes in the gene bank, designing the primers and probes, carrying out PCR (polymerase chain reaction) twice, carrying out molecular hybridization, and carrying out detection by using a Luminex200 system, thereby determining whether the sample contains the drug-resistant mutation sites. The detection of drug-resistant gene mutation sites is of crucial importance for treating Mycobacterium tuberculosis infection by adopting correct therapeutic schedules. The primers, probes and method have the advantages of high detection speed, high sensitivity, high specificity and the like, are simple to operate, and are beneficial to popularization and application.
Owner:HAINAN MEDICAL COLLEGE +1
Method for screening megalobrama amblycephala group combinations with hybrid vigor
InactiveCN102487860AObvious hybrid vigorImprove the breeding effectMicrobiological testing/measurementClimate change adaptationGene typeGenotype
The invention discloses a method for screening megalobrama amblycephala group combinations with hybrid vigor. The method comprises the following steps of: a, collecting wild parents of the megalobrama amblycephala in an original production area, and matching and breeding; b, culturing filial generations in the same pond in a mixed manner, and measuring relevant growth characteristics of the filial generations after 18 months; c, identifying the source of parents of the filial generations based on microsatellite markers; and d, based on a paternity test and growth characteristic data, analyzing the group combinations with the hybrid vigor, and finding that weights, lengths and heights of the filial generations in three combinations are obviously higher than that of other combinations. Different sources of wild parents are subjected to group combination breeding; a parent gene pool is enlarged; more gene types are collected; the hybrid vigor is sufficiently utilized; and the filial generations grow faster, thus, the megalobrama amblycephala selective breeding effect is more obvious.
Owner:HUAZHONG AGRI UNIV
Protein coded sequence for regulating and controlling temperature and light sensitive nuclear sterility
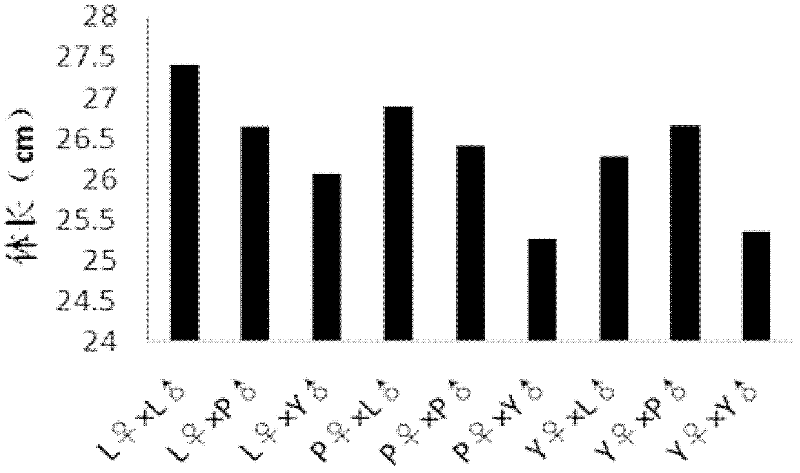
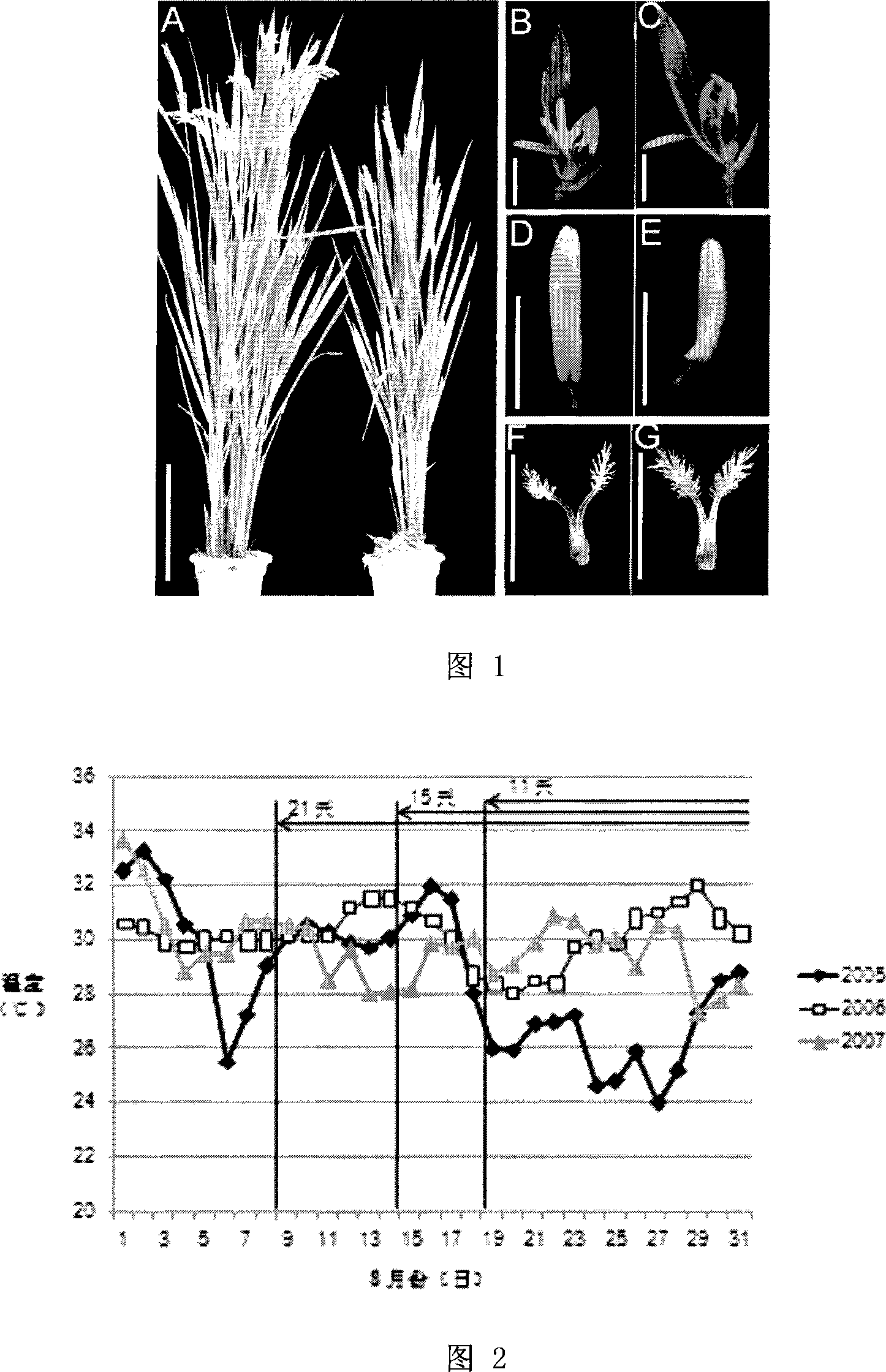
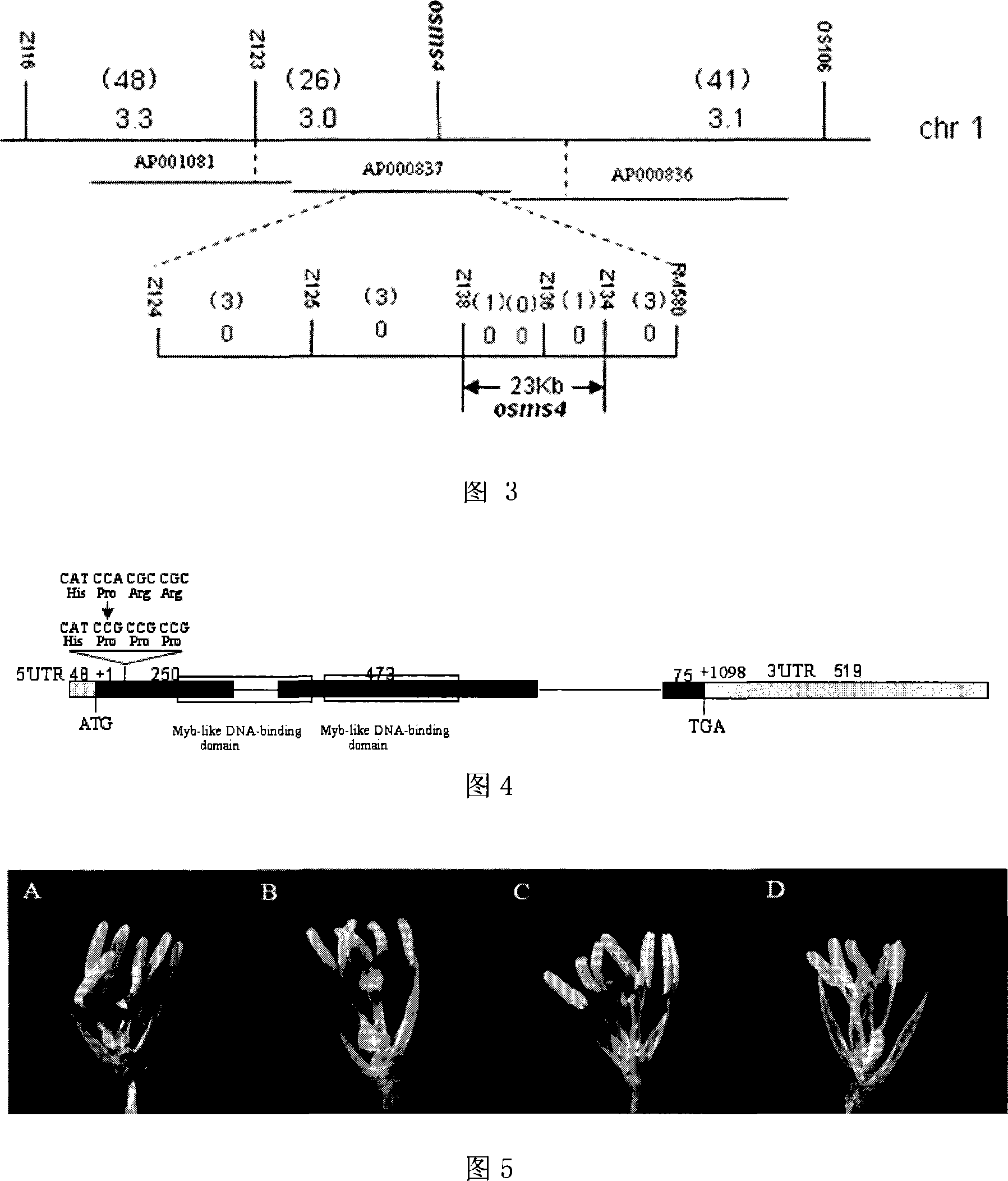
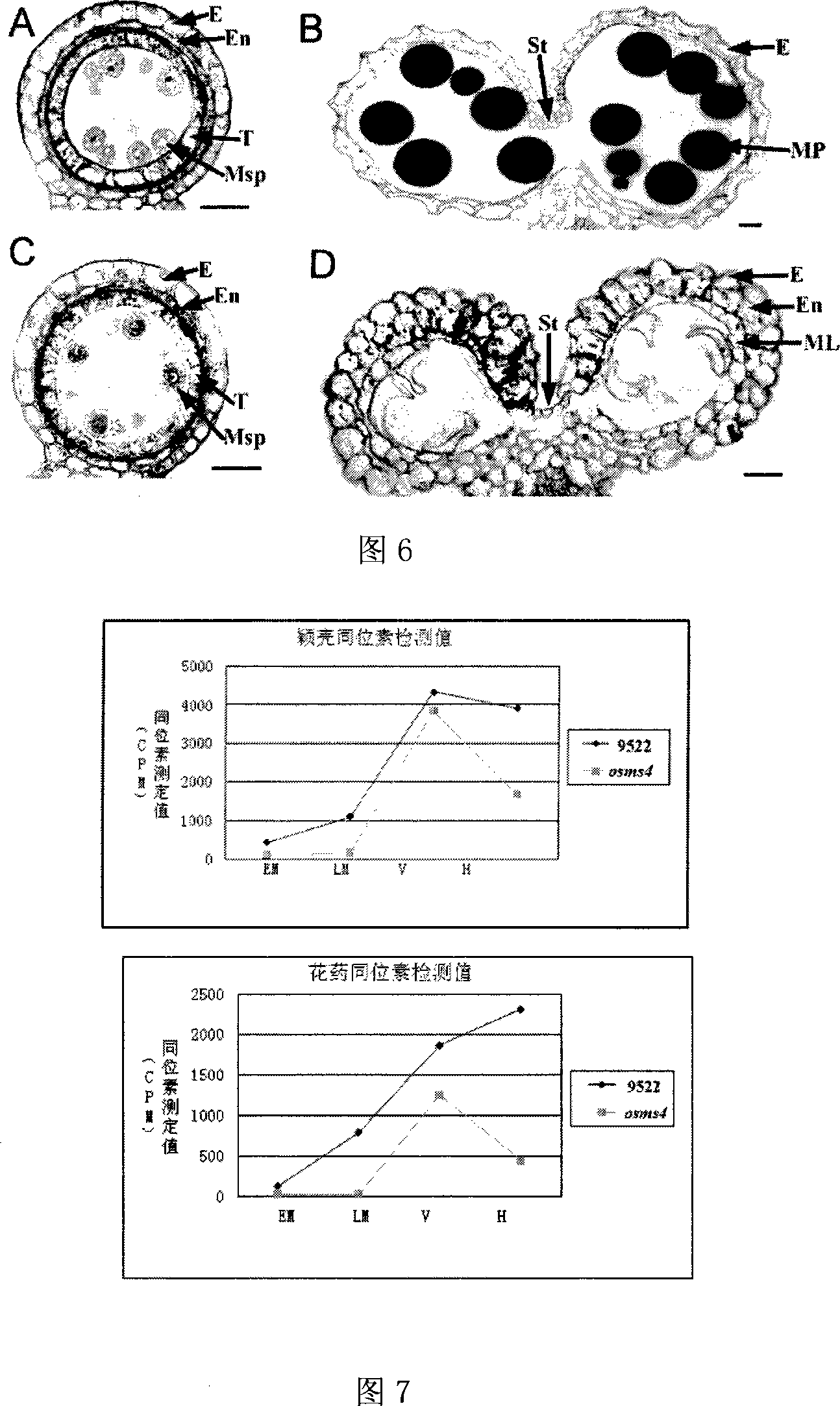
Provided is a protein coding sequence of regulating thermo-photo sensitive male sterile, which belongs to the field of gene engineering. The invention discloses a separated DNA molecule, including poly peptide nucleotide sequence with OsMS4 protein activation of rice of the code. The sequence has peptide of the amino acid sequence shown by the SEQ ID NO.2 and the nucleotide sequence from 1 nt to 798 nt shown by the SEQ ID NO.1. The invention has evident effect on detecting and utilizing rice thermo-photo sensitive male sterile. The invention can generate new male sterile line, which is used to produce hybrid, develop new type of two-line hybrid rice, select recurrently and create gene pool. Thereby the invention has a very important application to agricultural production.
Owner:上海旗冰种业科技有限公司
HPV16E7 protein nano-antibody as well as preparation method and application thereof
ActiveCN106397582AQuality improvementBiological material analysisImmunoglobulins against virusesEscherichia coliAntigen
The invention discloses an HPV16E7 protein nano-antibody as well as a preparation method and application thereof. According to the preparation method, camels are immunized by virtue of an HPV16E7 antigen expressed in a prokaryotic manner so as to obtain a high-quality immune nano-antibody gene pool; and an enzyme label plate is coated with HPV16E7 proteins, the immune nano-antibody gene pool is screened by virtue of a phage display technique to obtain nano-antibody genes with the HPV16E7 specificity, and then the nano-antibody genes are transferred to escherichia coli, so as to establish nano-antibody strains capable of realizing high-efficiency expression in the escherichia coli.
Owner:SOUTHEAST UNIV
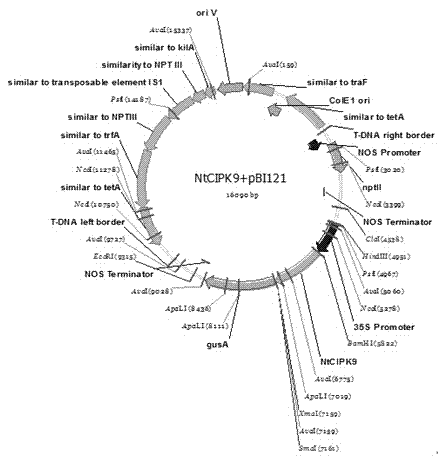
Nitraria tangutorum CBL-interacting protein kinase 9 (NtCIPK9) gene, expressed protein thereof and application thereof
ActiveCN104498514AImprove salt tolerance researchIncrease resourcesBacteriaTransferasesNucleotideAmino acid
The invention discloses a nitraria tangutorum CBL-interacting protein kinase 9 (NtCIPK9) gene, expressed protein of the NtCIPK9 gene and application of the NtCIPK9 gene. The nucleotide sequence of the NtCIPK9 gene is shown as the SEQ ID NO.1, and the amino acid sequence of the NtCIPK9 gene is shown as the SEQ ID NO.2. According to the NtCIPK9 gene, a full-length gene related to stress resistance of nitraria tangutorum is cloned in a homological mode by using leaf blade tissue of the nitraria tangutorum on the basis of a part of existing transcriptome data according to the stress resistance characteristic of the nitraria tangutorum, and is named NtCIPK9 according to a homologous gene in arabidopsis. The function of plant salt tolerance of the NtCIPK9 gene is proved through analysis of salt tolerance of an NtCIPK9 gene homozygous arabidopsis plant, and resources are increased for a plant stress resistance gene pool.
Owner:NANJING FORESTRY UNIV
Malware detection method based on software genetic technology
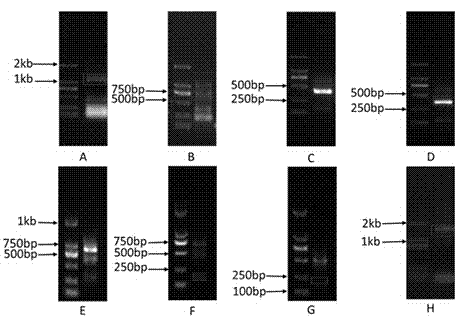
The invention discloses a malware detection method based on software genetic technology. The method comprises: performs gene extraction on a to-be-detected sample, comparing an extracted genome with azombie-trojan-worm virus family gene pool, calculating similarity, thereby identifying whether the to-be-detected sample is malware or not. The method can be used as an auxiliary detection means of malware, and effectively reduces missing report rate of malware detection by an existing malware detection method.
Owner:PEKING UNIV
Evolutionary clustering algorithm
ActiveUS20120016826A1Easy searchImprove the search of findingGenetic modelsDigital computer detailsCandidate Gene Association StudyGene pool
The invention relates to selecting a set of candidate genes from a pool of genes. The method comprising receiving a set of gene data; arranging the set of gene data into a set of clusters with similar profiles by use of a clustering algorithm; and inputting the set of clusters into a genetic algorithm to select a set of candidate genes from the set of clusters. The method thus relates to hybrid between selection by clustering computation and selection by evolutionary computation. This hybrid is also referred to as an evolutionary clustering algorithm (ECA).
Owner:KONINKLIJKE PHILIPS ELECTRONICS NV
Molecular genetic breeding method for California weever
InactiveCN108552088AGood characterClear genetic backgroundClimate change adaptationAnimal feeding stuffBroodstockEnvironment of Albania
The invention discloses a molecular genetic breeding method for California weever. The method comprises the steps of selecting basis parent fishes, establishing families, carrying out group breeding selection based on production cells, selecting and breeding parent fishes in the second generation families for hybridization, and repeating the above steps to obtain the third generation fishes. (1) The breeding method can efficiently breed a group with better characters, clear genetic backgrounds, larger gene pool and stable inheritance; (2) the breeding method can quickly screen parents with better characters to solve the problem of inbreeding depression; (3) the breeding method can directly breed fishes and select breeding in a large water body and directly carry out breeding selection under the environmental condition of production breeding, in this way, the result of breeding selection is more representative, the influence of environmental genes on fish growth can be further exerted so that genes of the breeding selection can make complete expression, in addition, the purpose that scientific research is combined with production is achieved, and when the breeding selection is carried out, production can be optimized, thereby acquiring a win-win effect.
Owner:NANJING SHUAIFENG FEED
Mycobacterium tuberculosis complex rapid detection kit and preparation method thereof
PendingCN111269996AQuick screeningTo achieve the purpose of comprehensive inspectionMicrobiological testing/measurementMicroorganism based processesSensitivity testMycobacterium
The invention discloses a mycobacterium tuberculosis complex rapid detection kit, which comprises an identification reagent, and the identification reagent comprises at least one pair of ten pairs ofprimers and at least one of ten probes. The invention also provides a preparation method of the rapid detection kit for the mycobacterium tuberculosis complex. The preparation method comprises the following steps of: S1, designing primers and probes according to gene sequences of the mycobacterium tuberculosis complex compared in a gene pool; S2, constructing a kit program of THE mycobacterium tuberculosis complex; S3, testing the specificity and sensitivity of the kit program; S4, carrying out repeated tests on the established kit program for many times; S5, preparing a standard target gene pool; and S6, constructing a specific detection kit with high sensitivity. The invention has the beneficial effects that by utilizing the kit based on the specific primer pair and the probe provided bythe invention, mycobacterium tuberculosis can be rapidly and comprehensively detected and identified, and possibility is provided for subsequent comprehensive detection; the sensitivity test, the specificity test and the repeatability test are utilized, and meanwhile, the CT difference value is compared, so that rapid and accurate detection is achieved.
Owner:WEST CHINA HOSPITAL SICHUAN UNIV +1
Method and System for Self-Optimized Uplink Power Control
A method for optimizing uplink power control settings in a wireless network, the method comprising generating a first gene pool comprising a set of parent genes, wherein each parent gene comprises a set of first generation power control solutions for a set of base stations in the wireless network. The method may further include performing natural selection on the first gene pool to generate a second gene pool comprising selected ones of the set of parent genes, wherein the selected parent genes are chosen by probabilistically selecting some of the parent genes based on fitness values assigned to the parent genes. The method may further include evolving the second gene pool into a descendent gene, wherein the descendent gene comprises a set of local power control solutions for the set of base station in the wireless network.
Owner:HONOR DEVICE CO LTD
Ammopiptanthus mongolicus cold-resistant gene AmEBP1
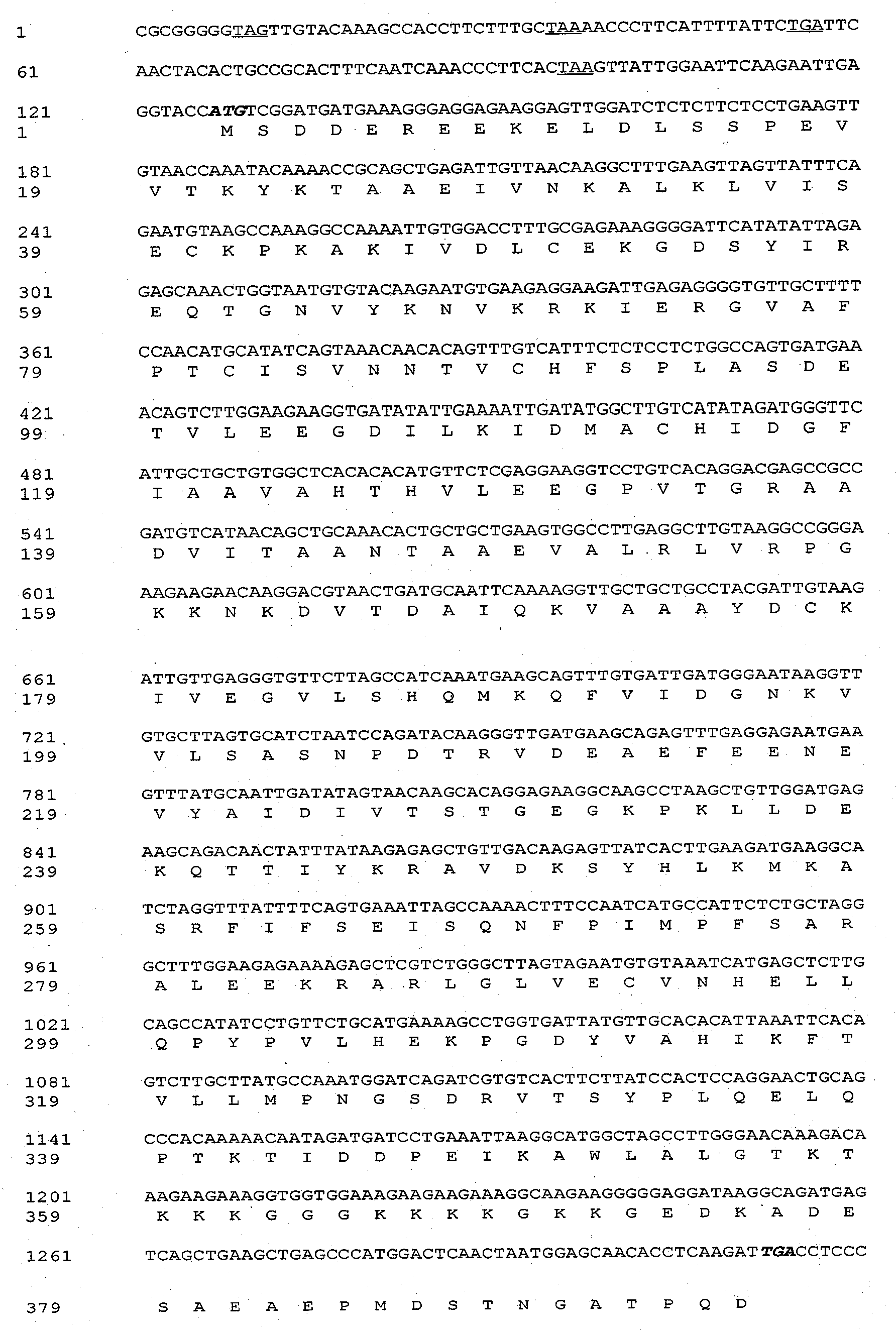
InactiveCN101892242AImprove cold resistanceGrowth inhibitionMicrobiological testing/measurementFermentationNucleotideTransgene
The invention discloses an ammopiptanthus mongolicus cold-resistant gene AmEBP1, and relates to a national secondary endangered and protected evergreen broad-leaved woody plant derived from the tertiary period in the desert regions of northwest China. The molecular cloning of the gene comprises the following steps of: obtaining a low temperature-induced and up regulation-expressed EST fragment through low temperature induction and differential expression analysis; and obtaining a full-length cDNA sequence of the gene through RACE amplification, and registering in a gene bank with the registration number of DQ519359. The AmEBP1 contains a whole ORF region with 1,188 nucleotides at the overall length, and the nucleotides code 395 amino acids and a terminator codon (TGA); a eukaryotic expression vector pCAMBIA2300-AmEBR1 is used to transform arabidopsis thaliana, and the obtained transgenic plant has improved cold resistance; and the ammopiptanthus mongolicus cold-resistant gene AmEBP1 is the first international woody plant cold-resistant gene which is cloned and passes genetic transformation verification.
Owner:TAISHAN RES INST OF FORESTRY +1
Molecular marker suitable for detecting radish leaf margin lobe trait and application of molecular marker
ActiveCN104313149AStabilized dominant SCAR markersSimple Assisted Breeding MethodMicrobiological testing/measurementDNA preparationAgricultural sciencePhenotypic trait
The invention belongs to the technical field of preparation of molecular markers of vegetables and particularly relates to a molecular marker suitable for detecting the radish leaf margin lobe trait and an application of the molecular marker. A molecular marker HY-18 which is tightly interlocked with a radish leaf margin lobe gene is obtained through the steps of selecting an appropriate parent to hybridize to obtain a plant F1; seeding a seed F1, bagging the plant F1 to inbreed and backcross to obtain segregation populations F2 and BC1; constructing a radish leaf supersulcus gene pool and a total leaf lobe gene pool, designing a specific primer, carrying out PCR, recovering, cloning, sequencing and screening; and the nucleotide sequence of the molecular marker is as shown in SEQ ID NO.1 in a sequence table. The invention also discloses a preparation method of the molecular marker. The molecular marker prepared by using the preparation method can be used for analyzing radish leaf lobe populations, and the practical marker and method are provided for molecular breeding of radishes.
Owner:WUHAN SHUBO AGRI TECH CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com